Abstract
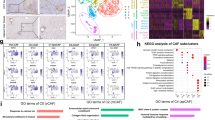
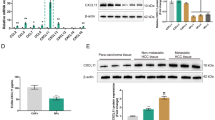
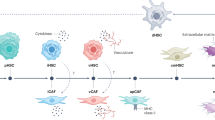
Cancer-associated fibroblasts (CAF) are important constituents of the tumor microenvironment (TME) and are major drivers of tumorigenesis. Yet, therapies aiming at eliminating CAF have failed to cure patients. This setback has raised questions regarding whether CAF exclusively favour cancer progression, or if they may also assume tumor-suppressor functions. In the present study, we used proteomics and single cell RNA-sequencing analysis to examine the CAF landscape in hepatocellular carcinoma (HCC). We thereby unveil three major CAF populations in HCC, one of which specifically expressing the prolargin protein. This CAF subpopulation (further termed as CAF_Port) shared a strong transcriptomic signature with portal liver fibroblasts. We further show that CAF_Port deposit prolargin in the TME and that its levels are lower in tumors as compared to the peritumoral region. Mechanistically, aggressive cancer cells degraded prolargin using matrix metalloprotease activity. Survival analysis of 188 patients revealed that high prolargin protein levels correlate with good patient outcome (HR = 0.37; p = 0.01). In vivo, co-injection of cancer cells with fibroblasts silenced for prolargin, led to faster tumor development (5-fold; p = 0.01), mainly due to stronger angiogenesis. Using protein-protein interaction study and structural modelling, we further demonstrate that prolargin binds and inhibits the activity of several pro-agiogenic proteins, including hepatocyte and fibroblast growth factors. In conclusion, prolargin is angiogenesis modulator and CAF-derived tumor suppressor in HCC. Stabilizing prolargin levels in the CAF_Port subpopulation may revert their tumor-antagonizing properties, warranting exploration in further pre-clinical studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76.
Wong MC, Jiang JY, Goggins WB, Liang M, Fang Y, Fung FDH, et al. International incidence and mortality trends of liver cancer: a global profile. Sci Rep. 2017;7:45846.
Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–91.
Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, et al. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:912–20.
Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl J Med. 2008;359:378–90.
Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl J Med. 2020;382:1894–905.
Voron T, Colussi O, Marcheteau E, Pernot S, Nizard M, Pointet AL, et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med. 2015;212:139–48.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37.
Ronca R, Van Ginderachter JA, Turtoi A. Paracrine interactions of cancer-associated fibroblasts, macrophages and endothelial cells: Tumor allies and foes. Curr Opin Oncol. 2018;30:45–53.
Chiavarina B, Turtoi A. Collaborative and defensive fibroblasts in tumor progression and therapy resistance. Curr Med Chem. 2017;24:2846–59.
Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61.
Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29.
Catenacci DV, Junttila MR, Karrison T, Bahary N, Horiba MN, Nattam SR, et al. Randomized Phase Ib/II Study of Gemcitabine Plus Placebo or Vismodegib, a Hedgehog pathway inhibitor, in patients with metastatic pancreatic cancer. J Clin Oncol. 2015;33:4284–92.
Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34.
Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47.
Dolberg DS, Bissell MJ. Inability of Rous sarcoma virus to cause sarcomas in the avian embryo. Nature. 1984;309:552–6.
Dotto GP, Weinberg RA, Ariza A. Malignant transformation of mouse primary keratinocytes by Harvey sarcoma virus and its modulation by surrounding normal cells. Proc Natl Acad Sci USA. 1988;85:6389–93.
Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–9.
Froeling FE, Feig C, Chelala C, Dobson R, Mein CE, Tuveson DA, et al. Retinoic acid-induced pancreatic stellate cell quiescence reduces paracrine Wnt-beta-catenin signaling to slow tumor progression. Gastroenterology. 2011;141:1486–97. 1497 e1–14.
Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93.
Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–96.
Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–79 e10.
Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172:841–56 e16.
Chang PH, Hwang-Verslues WW, Chang YC, Chen CC, Hsiao M, Jeng YM, et al. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/beta-catenin pathway. Cancer Res. 2012;72:4652–61.
DeFilippis RA, Chang H, Dumont N, Rabban JT, Chen YY, Fontenay GV, et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Disco. 2012;2:826–39.
Neill T, Schaefer L, Iozzo RV. Decorin: A guardian from the matrix. Am J Pathol. 2012;181:380–7.
Maris P, Blomme A, Palacios AP, Costanza B, Bellahcène A, Bianchi E, et al. Asporin Is a Fibroblast-Derived TGF-beta1 inhibitor and a tumor suppressor associated with good prognosis in breast cancer. PLoS Med. 2015;12:e1001871.
Turtoi A, Dumont B, Greffe Y, Blomme A, Mazzucchelli G, Delvenne P, et al. Novel comprehensive approach for accessible biomarker identification and absolute quantification from precious human tissues. J Proteome Res. 2011;10:3160–82.
Turtoi A, De Pauw E, Castronovo V. Innovative proteomics for the discovery of systemically accessible cancer biomarkers suitable for imaging and targeted therapies. Am J Pathol. 2011;178:12–8.
Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36:418–.e6.
Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–.e8.
Cabello-Aguilar S, Alame M, Kon-Sun-Tack F, Fau C, Lacroix M, Colinge J. SingleCellSignalR: Inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020;48:e55.
Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–50.
Rämisch S, Weininger U, Martinsson J, Akke M, André I. Computational design of a leucine-rich repeat protein with a predefined geometry. Proc Natl Acad Sci USA. 2014;111:17875–80.
Adnane L, Trail PA, Taylor I, Wilhelm SM. Sorafenib (BAY 43-9006, Nexavar), A dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Meth Enzymol. 2006;407:597–612.
Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7:3129–40.
Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–26.
Orimo A, Weinberg RA. Heterogeneity of stromal fibroblasts in tumors. Cancer Biol Ther. 2007;6:618–9.
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708–18.
Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhibitors in cancer therapy: Turning past failures into future successes. Mol Cancer Ther. 2018;17:1147–55.
Fields GB. Mechanisms of action of novel drugs targeting angiogenesis-promoting matrix metalloproteinases. Front Immunol. 2019;10:1278.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–21.
Waltregny D, Bellahcene A, Van Riet I, Fisher LW, Young M, Fernandez P, et al. Prognostic value of bone sialoprotein expression in clinically localized human prostate cancer. J Natl Cancer Inst. 1998;90:1000–8.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412.
Nwosu ZC, Battello N, Rothley M, Piorońska W, Sitek B, Ebert MP, et al. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J Exp Clin Cancer Res. 2018;37:211.
Acknowledgements
The authors acknowledge the experimental support of Naima Maloujahmoum (Metastasis Research Laboratory, University of Liege), Susumu Rokudai (Department of Molecular Pharmacology and Oncology, Gunma University), and Tadashi Handa (Pathology Dept. Gunma University). The authors are particularly thankful to Arnaud Blomme (Metastasis Research Laboratory, University of Liege) and Touko Hirano (Laboratory for Analytical Instruments, Gunma University Graduate School of Medicine) for the help concerning the MS analysis of patient material. The authors thank the Small Animal Imaging Platform of Montpellier (IPAM, http://www.ipam.cnrs.fr/) for the help with animal experiments. AT is thankful to Jacques Colinge (Bioinformatics and Systems Biology group, IRCM, Montpellier) for his R teachings and to Peter Friedl (The University of Texas MD Anderson Cancer Center, Houston, Texas, USA) for the helpful discussions. This work was supported with grants from the University of Liège, National Fund for Scientific Research (FNRS), Gunma University (GIAR Research Program for Omics-Based Medical Science). BC is supported by a Fondation de France grant (No. 00078461). RR is supported by Associazione Italiana per la Ricerca sul Cancro (AIRC), grant number MFAG 18459 and IG 2019 - ID. 23151; SR is supported by Fondazione Umberto Veronesi fellowship. AT is a senior research fellow of the French National Institute of Health and Medical Research (INSERM) and is supported by LabEx MabImprove Starting Grant. OD is supported by a grant from the “Fondation Contre le Cancer”. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Author information
Authors and Affiliations
Contributions
Study concept and design: AT and MN; drafting of the paper: AT, MN, BC; data acquisition and evaluation: BC, RR, YO, SR, TY, PC, RS, and ET; bioinformatics analysis: AT; structure analysis and docking: RBS; statistical analysis: BC, RR, YO; provided human clinical samples: SG, OD, TO, and PD; evaluation of pathology/histology: DP, GF, LK, TO, and PD; critical revision of the paper for important intellectual content: AB, AM, VC, MN, and AT; obtained funding: VC, MN, and AT; study supervision: AT.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chiavarina, B., Ronca, R., Otaka, Y. et al. Fibroblast-derived prolargin is a tumor suppressor in hepatocellular carcinoma. Oncogene 41, 1410–1420 (2022). https://doi.org/10.1038/s41388-021-02171-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02171-z
This article is cited by
-
CAFs vs. TECs: when blood feuds fuel cancer progression, dissemination and therapeutic resistance
Cellular Oncology (2024)
-
Correlation of the tumor escape phenotype with loss of PRELP expression in melanoma
Journal of Translational Medicine (2023)
-
Friend or foe? The elusive role of hepatic stellate cells in liver cancer
Nature Reviews Gastroenterology & Hepatology (2023)
-
Advances of cancer-associated fibroblasts in liver cancer
Biomarker Research (2022)