Abstract
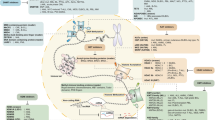
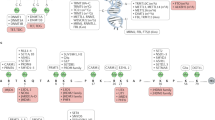
Hematological malignancies are a class of malignant neoplasms attributed to abnormal differentiation of hematopoietic stem cells (HSCs). The systemic involvement, poor prognosis, chemotherapy resistance, and recurrence common in hematological malignancies urge researchers to look for novel treatment targets and mechanisms. In recent years, epigenetic abnormalities have been shown to play a vital role in tumorigenesis and progression in hematological malignancies. In addition to DNA methylation and histone modifications, which are most studied, RNA methylation has become increasingly significant. In this review, we elaborate recent advances in the understanding of RNA modification in the pathogenesis, diagnosis and molecular targeted therapies of hematological malignancies and discuss its intricate interactions with other epigenetic modifications, including DNA methylation, histone modifications and noncoding RNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Delia D, Mizutani S. The DNA damage response pathway in normal hematopoiesis and malignancies. Int J Hematol. 2017;106:328–34.
Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30:2021–41.
Mendez-Ferrer S, Bonnet D, Steensma DP, Hasserjian RP, Ghobrial IM, Gribben JG, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer. 2020;20:285–98.
Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41.
Dimopoulos K, Gronbaek K. Epigenetic therapy in hematological cancers. APMIS. 2019;127:316–28.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7.
Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50.
Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci USA. 2018;115:E325–33.
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, et al. Regulation of Co-transcriptional Pre-mRNA splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell. 2019;76:70–81.e9.
Roundtree IA, Luo GZ, Zhang ZJ, Wang X, Zhou T, Cui YQ, et al. YTHDC1 mediates nuclear export of N-6 - methyladenosine methylated mRNAs. Elife 2017;6:e31311.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46.
Tuck MT. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int J Biochem. 1992;24:379–86.
Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47.
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–8.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna. 2018;24:499–512.
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73.
Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 2015;12:562–72.
Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–35.e14.
Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, et al. Human METTL16 is a N-6-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. Embo Rep. 2017;18:2004–14.
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10.
Wen J, Lv RT, Ma HH, Shen HJ, He CX, Wang JH, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028–38.e6.
Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m(6)A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. N Phytol. 2017;215:157–72.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Zhang X, Wei LH, Wang Y, Xiao Y, Liu J, Zhang W, et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci USA. 2019;116:2919–24.
Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, et al. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–83.
Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature. 2017;541:371–5.
Mauer J, Jaffrey SR. FTO, m(6) Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018;592:2012–22.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A reader YTHDC1 Regulates mRNA splicing. Mol Cell. 2016;61:507–19.
Du H, Zhao Y, He JQ, Zhang Y, Xi HR, Liu MF, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Alarcon CR, Goodarzi H, Lee H, Liu XH, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–308.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan TN. (6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4.
Zheng W, Dong X, Zhao Y, Wang S, Jiang H, Zhang M, et al. Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol. 2019;9:1403.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Lee H, Bao SY, Qian YZ, Geula S, Leslie J, Zhang CL, et al. Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21:700–9.
Cheng Y, Luo H, Izzo F, Pickering BF, Nguyen D, Myers R, et al. m(6)A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 2019;28:1703–16.e6.
Sheng Y, Ma R, Yu C, Wu Q, Zhang S, Paulsen K, et al. Role of c-Myc haploinsufficiency in the maintenance of HSCs in mice. Blood 2021;137:610–23.
Yao QJ, Sang L, Lin M, Yin X, Dong W, Gong Y, et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 2018;28:952–4.
Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–31.
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60.
Albertella M, Blackaby W, Fosbeary R, Hendrick A, Leggate D, Ofir-Rosenfeld Y, et al. A small molecule inhibitor of the RNA m6A writer METTL3 inhibits the development of acute myeloid leukemia (AML) in vivo. Mol Cancer Ther 2019;18(12 Supplement):B126–B126.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43.
Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol Cancer. 2019;18:168.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e9.
Sun C, Chang L, Liu C, Chen X, Zhu X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Mol Genet Genom Med. 2019;7:e00933.
Bansal H, Yihua Q, Iyer SP, Ganapathy S, Proia D, Penalva LO, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–4.
Sorci M, Ianniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis 2018;9:796.
Wiseman DH, Bonney DK, Wynn RF. Hemophagocytosis by leukemic megakaryoblasts in acute myeloid leukemia (Megakaryoblastic) With t(1;22)(p13;q13);RBM15-MKL1. J Pediatr Hematol Oncol. 2012;34:576–80.
Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–64.
Schuschel K, Helwig M, Huttelmaier S, Heckl D, Klusmann JH, Hoell JI. RNA-Binding Proteins in Acute Leukemias. Int J Mol Sci 2020;21:3409.
Jin S, Mi Y, Song J, Zhang P, Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34(+) cells. Exp Ther Med. 2018;15:2563–8.
Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang H, et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. 2015;4:e07938.
Tran NT, Su HR, Khodadadi-Jamayran A, Lin S, Zhang L, Zhou DW, et al. The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. Embo Rep. 2016;17:887–900.
Deng XL, Su R, Stanford S, Chen JJ. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol. 2018;9:396.
Li ZJ, Weng HY, Su R, Weng XC, Zuo ZX, Li CY, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N-6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34.
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46.
Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019;145:19–29.
Yan F, Al-Kali A, Zhang Z, Liu J, Pang J, Zhao N, et al. A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28:1062–76.
Xu A, Zhang J, Zuo L, Xu J, Yin X, Cheng Q, et al. Fto promotes extramedullary progression of multiple myeloma by regulation of HSF1 through m6a RNA methylation. Blood. 2019;134:3063.
Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134:17963–71.
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e123.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–91.e10.
Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96.e11.
Zhang CZ, Samanta D, Lu HQ, Bullen JW, Zhang HM, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6.
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3.
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27:64–80.e9.
Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, et al. Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 2020;27:81–97 e88.
Li Q, Ye L, Guo W, Wang M, Huang S, Peng X. Overexpression of TACC3 is correlated with tumor aggressiveness and poor prognosis in prostate cancer. Biochem Biophys Res Commun. 2017;486:872–8.
Lin ZR, Wang MY, He SY, Cai ZM, Huang WR. TACC3 transcriptionally upregulates E2F1 to promote cell growth and confer sensitivity to cisplatin in bladder cancer. Cell Death Dis. 2018;9:72.
Moritsubo M, Miyoshi H, Matsuda K, Yoshida N, Nakashima K, Yanagida E, et al. TACC3 expression as a prognostic factor in aggressive types of adult T-cell leukemia/lymphoma patients. Int J Lab Hematol 2020;42:842–48.
Shen DD, Suo FZ, Song QM, Chang J, Zhang T, Hong JJ, et al. Development of formaldehyde dehydrogenase-coupled assay and antibody-based assays for ALKBH5 activity evaluation. J Pharm Biomed Anal. 2019;162:9–15.
Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg Med Chem. 2020;28:115300.
Stoskus M, Gineikiene E, Valceckiene V, Valatkaite B, Pileckyte R, Griskevicius L. Identification of characteristic IGF2BP expression patterns in distinct B-ALL entities. Blood Cells Mol Dis. 2011;46:321–6.
Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2020;34:1354–63.
Zhou J, Bi C, Ching YQ, Chooi JY, Lu X, Quah JY, et al. Inhibition of LIN28B impairs leukemia cell growth and metabolism in acute myeloid leukemia. J Hematol Oncol. 2017;10:138.
Vijayakrishnan J, Qian M, Studd JB, Yang W, Kinnersley B, Law PJ, et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat Commun. 2019;10:5348.
Jeffries SJ, Jones L, Harrison CJ, Russell LJ. IGH@ translocations co-exist with other primary rearrangements in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2014;99:1334–42.
Gu G, Sederberg MC, Drachenberg MR, South ST. IGF2BP1: a novel IGH translocation partner in B acute lymphoblastic leukemia. Cancer Genet. 2014;207:332–4.
Stoskus M, Eidukaite A, Griskevicius L. Defining the significance of IGF2BP1 overexpression in t(12;21)(p13;q22)-positive leukemia REH cells. Leuk Res. 2016;47:16–21.
Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36:498–511.e7.
He X, Li W, Liang X, Zhu X, Zhang L, Huang Y, et al. IGF2BP2 overexpression indicates poor survival in patients with acute myelocytic leukemia. Cell Physiol Biochem. 2018;51:1945–56.
Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Investig. 2016;126:1495–511.
Liao B, Hu Y, Brewer G. RNA-binding protein insulin-like growth factor mRNA-binding protein 3 (IMP-3) promotes cell survival via insulin-like growth factor II signaling after ionizing radiation. J Biol Chem. 2011;286:31145–52.
Ko CY, Wang WL, Li CF, Jeng YM, Chu YY, Wang HY, et al. IL-18-induced interaction between IMP3 and HuR contributes to COX-2 mRNA stabilization in acute myeloid leukemia. J Leukoc Biol. 2016;99:131–41.
Carter BZ, Mak PY, Wang X, Tao W, Ruvolo V, Mak D, et al. An ARC-regulated IL1beta/Cox-2/PGE2/beta-Catenin/ARC circuit controls leukemia-microenvironment interactions and confers drug resistance in AML. Cancer Res. 2019;79:1165–77.
Masoud R, Ibrahiem A, Tantawy D, Eldosoky I. The complementary role of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in diagnosis of Hodgkin’s lymphoma. Ann Diagn Pathol. 2019;42:64–8.
Li ZR, Qian PX, Shao WQ, Shi HL, He XC, Gogol M, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion (vol 28, pg 904, 2018). Cell Res. 2018;28:1042.
Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28:1035–8.
Mapperley C, van de Lagemaat LN, Lawson H, Tavosanis A, Paris J, Campos J, et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J Exp Med. 2021;218:e20200829.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–48.e6.
Furuya K, Ikura M, Ikura T. Epigenetic interplays between DNA demethylation and histone methylation for protecting oncogenesis. J Biochem. 2019;165:297–9.
Scalea S, Maresca C, Catalanotto C, Marino R, Cogoni C, Reale A, et al. Modifications of H3K4 methylation levels are associated with DNA hypermethylation in acute myeloid leukemia. Febs J. 2020;287:1155–75.
Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X, et al. m(6)A Regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom Proteom Bioinform. 2019;17:154–68.
Wang Y, Li Y, Yue MH, Wang J, Kumar S, Wechsler-Reya RJ, et al. N-6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206.
Lai WW, Jia JT, Yan B, Jiang YQ, Shi Y, Chen L, et al. Baicalin hydrate inhibits cancer progression in nasopharyngeal carcinoma by affecting genome instability and splicing. Oncotarget. 2018;9:901–14.
Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, et al. N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52:870–7.
Wu C, Chen W, He J, Jin S, Liu Y, Yi Y, et al. Interplay of m(6)A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci Adv. 2020;6:eaba0647.
Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–89.
Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414–9.
Zhang JL, Bai RH, Li M, Ye HL, Wu C, Wang CF, et al. Excessive miR-25-3p maturation via N-6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858.
Zhang J, Zheng YG. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. Acs Chem Biol. 2016;11:583–97.
Xiao CL, Zhu S, He M, Chen, Zhang Q, Chen Y, et al. N(6)-Methyladenine DNA modification in the human genome. Mol Cell. 2018;71:306–18.e7.
Lin SB, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244–55.e5.
Boulias K, Greer EL. Put the pedal to the METTL1: adding Internal m(7)G increases mRNA translation efficiency and augments miRNA processing. Mol Cell. 2019;74:1105–7.
Xu L, Liu XY, Sheng N, Oo KS, Liang JX, Chionh YH, et al. Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem. 2017;292:14695–703.
Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol Cell. 2020;78:382–95.e8.
Chen H, Gu L, Orellana EA, Wang Y, Guo J, Liu Q, et al. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020;30:544–47.
van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33.
Shimazu T, Barjau J, Sohtome Y, Sodeoka M, Shinkai Y. Selenium-based S-adenosylmethionine analog reveals the mammalian seven-beta-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. Plos ONE. 2014;9:e105394.
Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, et al. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210.
Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, Merchant ML, Macara IG, et al. NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. Biochem J. 2013;456:453–62.
Malecki J, Jakobsson ME, Ho AYY, Moen A, Rustan AC, Falnes PO. Uncovering human METTL12 as a mitochondrial methyltransferase that modulates citrate synthase activity through metabolite-sensitive lysine methylation. J Biol Chem. 2017;292:17950–62.
Jakobsson ME, Malecki JM, Halabelian L, Nilges BS, Pinto R, Kudithipudi S, et al. The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat Commun. 2018;9:3411.
Van Haute L, Hendrick AG, D’Souza AR, Powell CA, Rebelo-Guiomar P, Harbour ME, et al. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019;47:10267–81.
Shi ZN, Xu SY, Xing SH, Yao K, Zhang L, Xue LX, et al. Mettl17, a regulator of mitochondrial ribosomal RNA modifications, is required for the translation of mitochondrial coding genes. Faseb J. 2019;33:13040–50.
Malecki J, Ho AYY, Moen A, Dahl HA, Falnes PO. Human METTL20 is a mitochondrial lysine methyltransferase that targets the beta subunit of electron transfer flavoprotein (ETF beta) and modulates its activity. J Biol Chem. 2015;290:423–34.
Malecki J, Aileni VK, Ho AYY, Schwarz J, Moen A, Sorensen V, et al. The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 2017;45:4370–89.
Wiederstein JL, Nolte H, Gunther S, Piller T, Baraldo M, Kostin S, et al. Skeletal muscle-specific methyltransferase METTL21C trimethylates p97 and regulates autophagy-associated protein breakdown. Cell Rep. 2018;23:1342–56.
Wang C, Arrington J, Ratliff AC, Chen JJ, Horton HE, Nie YH, et al. Methyltransferase-like 21c methylates and stabilizes the heat shock protein Hspa8 in type I myofibers in mice. J Biol Chem. 2019;294:13718–28.
Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, et al. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038.
Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. Methylation of the DNA/RNA-binding protein Kin17 by METTL22 affects its association with chromatin. J Proteom. 2014;100:115–24.
Hatanaka Y, Tsusaka T, Shimizu N, Morita K, Suzuki T, Machida S, et al. Histone H3 methylated at arginine 17 is essential for reprogramming the paternal genome in zygotes. Cell Rep. 2017;20:2756–65.
Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33.
Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, et al. Structure of human RNA N(6)-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–54.
Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016;167:1897.
Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–56.
Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–19.
Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–98.
Fu D, Samson LD. Direct repair of 3,N-4-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair. 2012;11:46–52.
You CJ, Wang PC, Nay SL, Wang JS, Dai XX, O’Connor TR, et al. Roles of Aag, Alkbh2, and Alkbh3 in the repair of carboxymethylated and ethylated thymidine lesions. ACS Chem Biol. 2016;11:1332–8.
Bian K, Lenz SAP, Tang Q, Chen FY, Qi R, Jost M, et al. DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro. Nucleic Acids Res. 2019;47:5522–9.
Chen ZJ, Qi MJ, Shen B, Luo GZ, Wu YM, Li JX, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–45.
Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol. 2010;30:1814–27.
Martin Carli JF, LeDuc CA, Zhang Y, Stratigopoulos G, Leibel RL. FTO mediates cell-autonomous effects on adipogenesis and adipocyte lipid content by regulating gene expression via 6mA DNA modifications. J Lipid Res. 2018;59:1446–60.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020;21:1027.
Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–75.
Rodriguez-Malave NI, Rao DS. Long noncoding RNAs in hematopoietic malignancies. Brief Funct Genom. 2016;15:227–38.
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–26.
Zuo XL, Chen ZQ, Gao W, Zhang Y, Wang JG, Wang JF, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5.
Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020;12:4558–72.
Ban YY, Tan PQ, Cai J, Li JJ, Hu M, Zhou Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282–96.
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135.
Liu H, Xu Y, Yao B, Sui T, Lai L, Li Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613.
Yoneda R, Ueda N, Uranishi K, Hirasaki M, Kurokawa R. Long noncoding RNA pncRNA-D reduces cyclin D1 gene expression and arrests cell cycle through RNA m(6)A modification. J Biol Chem. 2020;295:5626–39.
Barros-Silva D, Lobo J, Guimaraes-Teixeira C, Carneiro I, Oliveira J, Martens-Uzunova ES, et al. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers. 2020;12:771.
Hu XG, Peng WX, Zhou HX, Jiang JH, Zhou XC, Huang DS, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–94.
Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34.
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48:838–46.
Yan JJ, Huang XF, Zhang XJ, Chen ZJ, Ye C, Xiang WC, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887–93.
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–72.
Lan T, Li H, Zhang DL, Xu L, Liu HL, Hao XY, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186.
Liu B, Zhou J, Wang C, Chi Y, Wei Q, Fu Z, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020;11:384.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21.
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N-6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110.
Peng W, Li J, Chen RR, Gu QO, Yang P, Qian WW, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393.
Wang HS, Deng QQ, Lv ZY, Ling YY, Hou X, Chen ZJ, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181.
Yi DD, Wang R, Shi XB, Xu L, Yilihamu Y, Sang JF. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol Rep. 2020;43:1375–86.
Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9:9430.
Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163.
Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han WF, et al. m(6)A RNA methylation is regulated by MicroRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301.
Cai XL, Wang X, Cao C, Gao YE, Zhang SQ, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768.
Shen XP, Ling X, Lu H, Zhou CX, Zhang JK, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2018;22:8220–6.
Xu C, Yuan B, He T, Ding B, Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J Cell Mol Med 2020;24:7538–49.
Yang Z, Li J, Feng GX, Gao S, Wang Y, Zhang SQ, et al. MicroRNA-145 modulates N-6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N-6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292:3614–23.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Di Timoteo G, Dattilo D, Centron-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31:107641.
Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507.e8.
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695.
Acknowledgements
This work is supported by the National Natural Science Foundation of PR China (no. 81570193 and no. 81770219, for QW).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yao, L., Yin, H., Hong, M. et al. RNA methylation in hematological malignancies and its interactions with other epigenetic modifications. Leukemia 35, 1243–1257 (2021). https://doi.org/10.1038/s41375-021-01225-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01225-1
This article is cited by
-
Impaired binding affinity of YTHDC1 with METTL3/METTL14 results in R-loop accumulation in myelodysplastic neoplasms with DDX41 mutation
Leukemia (2024)
-
SWI/SNF complexes in hematological malignancies: biological implications and therapeutic opportunities
Molecular Cancer (2023)
-
RNA modifications—a regulatory dimension yet to be deciphered in immunity
Genes & Immunity (2023)
-
The role of RNA modification in urological cancers: mechanisms and clinical potential
Discover Oncology (2023)
-
Evaluation of FTO polymorphism in 6-mercaptopurine related intolerance in children with acute lymphoblastic leukemia
Cancer Chemotherapy and Pharmacology (2023)