Abstract
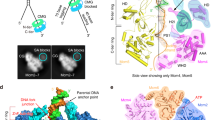
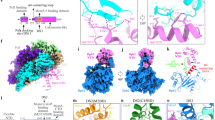
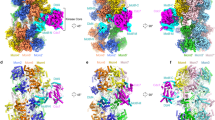
Two central steps for initiating eukaryotic DNA replication involve loading of the Mcm2–7 helicase onto double-stranded DNA and its activation by GINS–Cdc45. To better understand these events, we determined the structures of Mcm2–7 and the CMG complex by using single-particle electron microscopy. Mcm2–7 adopts two conformations—a lock-washer-shaped spiral state and a planar, gapped-ring form—in which Mcm2 and Mcm5 flank a breach in the helicase perimeter. GINS and Cdc45 bridge this gap, forming a topologically closed assembly with a large interior channel; nucleotide binding further seals off the discontinuity between Mcm2 and Mcm5, partitioning the channel into two smaller pores. Together, our data help explain how GINS and Cdc45 activate Mcm2–7, indicate that Mcm2–7 loading may be assisted by a natural predisposition of the hexamer to form open rings, and suggest a mechanism by which the CMG complex assists DNA strand separation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Davey, M.J. & O'Donnell, M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13, R594–R596 (2003).
Funnell, B.E., Baker, T.A. & Kornberg, A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 262, 10327–10334 (1987).
Davey, M.J., Fang, L., McInerney, P., Georgescu, R.E. & O'Donnell, M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 21, 3148–3159 (2002).
Valle, M., Gruss, C., Halmer, L., Carazo, J.M. & Donate, L.E. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20, 34–41 (2000).
Kumar, A. et al. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J. Virol. 81, 4808–4818 (2007).
Schuck, S. & Stenlund, A. Assembly of a double hexameric helicase. Mol. Cell 20, 377–389 (2005).
Enemark, E.J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006).
Bowers, J.L., Randell, J.C., Chen, S. & Bell, S.P. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol. Cell 16, 967–978 (2004).
Remus, D. et al. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 (2009).
Evrin, C. et al. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 106, 20240–20245 (2009).
Moyer, S.E., Lewis, P.W. & Botchan, M.R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103, 10236–10241 (2006).
Gambus, A. et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 (2006).
Remus, D. & Diffley, J.F. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21, 771–777 (2009).
Calzada, A., Hodgson, B., Kanemaki, M., Bueno, A. & Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19, 1905–1919 (2005).
Ilves, I., Petojevic, T., Pesavento, J.J. & Botchan, M.R. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 (2010).
Pacek, M., Tutter, A.V., Kubota, Y., Takisawa, H. & Walter, J.C. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 (2006).
MacNeill, S.A. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem. J. 425, 489–500 (2010).
Fletcher, R.J. et al. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat. Struct. Biol. 10, 160–167 (2003).
McGeoch, A.T., Trakselis, M.A., Laskey, R.A. & Bell, S.D. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 12, 756–762 (2005).
Iyer, L.M., Leipe, D.D., Koonin, E.V. & Aravind, L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 (2004).
Davey, M.J., Indiani, C. & O'Donnell, M. Reconstitution of the Mcm2–7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 278, 4491–4499 (2003).
Crevel, G., Ivetic, A., Ohno, K., Yamaguchi, M. & Cotterill, S. Nearest neighbour analysis of MCM protein complexes in Drosophila melanogaster. Nucleic Acids Res. 29, 4834–4842 (2001).
Bochman, M.L., Bell, S.P. & Schwacha, A. Subunit organization of Mcm2–7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell. Biol. 28, 5865–5873 (2008).
Pape, T. et al. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 4, 1079–1083 (2003).
Brewster, A.S. et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA 105, 20191–20196 (2008).
Bae, B. et al. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure 17, 211–222 (2009).
Yu, X. et al. The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep. 3, 792–797 (2002).
Gómez-Llorente, Y., Fletcher, R.J., Chen, X.S., Carazo, J.M. & San Martin, C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 280, 40909–40915 (2005).
Costa, A. et al. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 34, 5829–5838 (2006).
Chong, J.P., Hayashi, M.K., Simon, M.N., Xu, R.M. & Stillman, B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 97, 1530–1535 (2000).
Kelman, Z., Lee, J.K. & Hurwitz, J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl. Acad. Sci. USA 96, 14783–14788 (1999).
Bochman, M.L. & Schwacha, A. The Mcm2–7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 (2008).
Kanemaki, M., Sanchez-Diaz, A., Gambus, A. & Labib, K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423, 720–724 (2003).
Takayama, Y. et al. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17, 1153–1165 (2003).
Boskovic, J. et al. Molecular architecture of the human GINS complex. EMBO Rep. 8, 678–684 (2007).
Choi, J.M., Lim, H.S., Kim, J.J., Song, O.K. & Cho, Y. Crystal structure of the human GINS complex. Genes Dev. 21, 1316–1321 (2007).
Chang, Y.P., Wang, G., Bermudez, V., Hurwitz, J. & Chen, X.S. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. USA 104, 12685–12690 (2007).
Kamada, K., Kubota, Y., Arata, T., Shindo, Y. & Hanaoka, F. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat. Struct. Mol. Biol. 14, 388–396 (2007).
Grob, P. et al. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure 14, 511–520 (2006).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Im, J.S. et al. Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA 106, 15628–15632 (2009).
Makarova, K.S., Wolf, Y.I., Mekhedov, S.L., Mirkin, B.G. & Koonin, E.V. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33, 4626–4638 (2005).
Marinsek, N. et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7, 539–545 (2006).
Dean, F.B., Borowiec, J.A., Eki, T. & Hurwitz, J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267, 14129–14137 (1992).
Fouts, E.T., Yu, X., Egelman, E.H. & Botchan, M.R. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274, 4447–4458 (1999).
Bochman, M.L. & Schwacha, A. Differences in the single-stranded DNA binding activities of MCM2–7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 282, 33795–33804 (2007).
Labib, K. & Gambus, A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 17, 271–278 (2007).
Takahashi, T.S., Wigley, D.B. & Walter, J.C. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 30, 437–444 (2005).
Takara, T.J. & Bell, S.P. Putting two heads together to unwind DNA. Cell 139, 652–654 (2009).
Yardimci, H., Loveland, A.B., Habuchi, S., van Oijen, A.M. & Walter, J.C. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell 40, 834–840 (2010).
Gould, A.D. & Shilton, B.H. Studies of the maltose transport system reveal a mechanism for coupling ATP hydrolysis to substrate translocation without direct recognition of substrate. J. Biol. Chem. 285, 11290–11296 (2010).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Voss, N.R., Yoshioka, C.K., Radermacher, M., Potter, C.S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
van Heel, M. et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 (2000).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Boekema, E.J., Berden, J.A. & van Heel, M.G. Structure of mitochondrial F1-ATPase studied by electron microscopy and image processing. Biochim. Biophys. Acta 851, 353–360 (1986).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004).
Acknowledgements
The authors would like to thank A. Lyubimov and F. Bleichert for comments and help with the manuscript; and G. Lander, P. Grob, R. Hannah, R. Hall, M. Cianfrocco and C. Ciferri for technical help. This work was supported by a European Molecular Biology Organization long-term postdoctoral fellowship (to A.C.), a PhD fellowship from the Boehringer Ingelheim Fonds (to T.P.), the Human Frontier Science Program (RPG0039, to E.N.), the National Institute of General Medical Sciences (GM071747, to J.M.B.) and the National Cancer Institute (CA R37-30490, to M.R.B.). E.N. is a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Contributions
A.C., I.I., M.R.B. and J.M.B. conceived the general ideas for this study. All authors planned experiments. A.C. did all electron microscopy single-particle reconstruction and molecular modeling supervised by J.M.B. and E.N. I.I., N.T. and T.P. did cloning, baculovirus construction and protein purification supervised by M.R.B. A.C., M.R.B. and J.M.B. wrote the manuscript. All authors provided editorial input.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 1436 kb)
Supplementary Movie 1
The Mcm2-7 complex morphing between a planar-notched and a spiral-lockwasher configuration. (MOV 4475 kb)
Supplementary Movie 2
The CMG complex morphing between the apo and nucleotide-bound state (MOV 3815 kb)
Rights and permissions
About this article
Cite this article
Costa, A., Ilves, I., Tamberg, N. et al. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18, 471–477 (2011). https://doi.org/10.1038/nsmb.2004
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2004
This article is cited by
-
Nucleotide binding halts diffusion of the eukaryotic replicative helicase during activation
Nature Communications (2023)
-
Relatively semi-conservative replication and a folded slippage model for short tandem repeats
BMC Genomics (2020)
-
Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks
Cellular and Molecular Life Sciences (2020)
-
DNA translocation mechanism of the MCM complex and implications for replication initiation
Nature Communications (2019)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)