Abstract
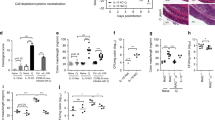
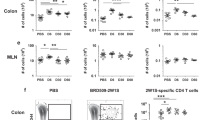
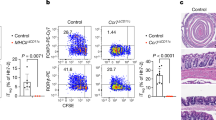
Interleukin 17 (IL-17) is a central cytokine implicated in inflammation and antimicrobial defense. After infection, both innate and adaptive IL-17 responses have been reported, but the type of cells involved in innate IL-17 induction, as well as their contribution to in vivo responses, are poorly understood. Here we found that Citrobacter and Salmonella infection triggered early IL-17 production, which was crucial for host defense and was mediated by CD4+ T helper cells. Enteric innate T helper type 17 (iTH17) responses occurred principally in the cecum, were dependent on the Nod-like receptors Nod1 and Nod2, required IL-6 induction and were associated with a decrease in mucosal CD103+ dendritic cells. Moreover, imprinting by the intestinal microbiota was fully required for the generation of iTH17 responses. Together, these results identify the Nod-iTH17 axis as a central element in controlling enteric pathogens, which may implicate Nod-driven iTH17 responses in the development of inflammatory bowel diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bettelli, E., Korn, T., Oukka, M. & Kuchroo, V.K. Induction and effector functions of TH17 cells. Nature 453, 1051–1057 (2008).
Ouyang, W., Kolls, J.K. & Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467 (2008).
Colonna, M. Interleukin-22–producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity 31, 15–23 (2009).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Veldhoen, M., Hocking, R.J., Atkins, C.J., Locksley, R.M. & Stockinger, B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17–producing T cells. Immunity 24, 179–189 (2006).
Littman, D.R. & Rudensky, A.Y. TH17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Takatori, H. et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 206, 35–41 (2009).
Billerbeck, E. et al. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc. Natl. Acad. Sci. USA 107, 3006–3011 (2010).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Martin, B., Hirota, K., Cua, D.J., Stockinger, B. & Veldhoen, M. Interleukin-17–producing γδ T cells selectively expand in response to pathogen products and environmental signals. Immunity 31, 321–330 (2009).
Sonnenberg, G.F., Monticelli, L.A., Elloso, M.M., Fouser, L.A. & Artis, D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011).
Aujla, S.J. et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14, 275–281 (2008).
Priebe, G.P. et al. IL-17 is a critical component of vaccine-induced protection against lung infection by lipopolysaccharide-heterologous strains of Pseudomonas aeruginosa. J. Immunol. 181, 4965–4975 (2008).
Khader, S.A. et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8, 369–377 (2007).
Umemura, M. et al. IL-17–mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J. Immunol. 178, 3786–3796 (2007).
Wu, Q. et al. IL-23–dependent IL-17 production is essential in neutrophil recruitment and activity in mouse lung defense against respiratory Mycoplasma pneumoniae infection. Microbes Infect. 9, 78–86 (2007).
Sellge, G. et al. TH17 cells are the dominant T cell subtype primed by Shigella flexneri mediating protective immunity. J. Immunol. 184, 2076–2085 (2010).
Velin, D. et al. Interleukin-17 is a critical mediator of vaccine-induced reduction of Helicobacter infection in the mouse model. Gastroenterology 136, 2237–2246 (2009).
Raffatellu, M. et al. Simian immunodeficiency virus–induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14, 421–428 (2008).
Raffatellu, M. et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009).
Ishigame, H. et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30, 108–119 (2009).
Godinez, I. et al. Interleukin-23 orchestrates mucosal responses to Salmonella enterica serotype Typhimurium in the intestine. Infect. Immun. 77, 387–398 (2009).
Symonds, E.L. et al. Involvement of T helper type 17 and regulatory T cell activity in Citrobacter rodentium invasion and inflammatory damage. Clin. Exp. Immunol. 157, 148–154 (2009).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Godinez, I. et al. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76, 2008–2017 (2008).
Geddes, K., Magalhaes, J.G. & Girardin, S.E. Unleashing the therapeutic potential of NOD-like receptors. Nat. Rev. Drug Discov. 8, 465–479 (2009).
LeBlanc, P.M. et al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe 3, 146–157 (2008).
Cash, H.L., Whitham, C.V., Behrendt, C.L. & Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Geddes, K. et al. Nod1 and Nod2 regulation of inflammation in the Salmonella colitis model. Infect. Immun. 78, 5107–5115 (2010).
Ivanov, I.I. et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Coombes, J.L. & Powrie, F. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 8, 435–446 (2008).
Niess, J.H. & Adler, G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J. Immunol. 184, 2026–2037 (2010).
Dutton, R.W., Bradley, L.M. & Swain, S.L. T cell memory. Annu. Rev. Immunol. 16, 201–223 (1998).
Ivanov, I.I. et al. Induction of intestinal TH17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Ivanov, I.I. et al. Specific microbiota direct the differentiation of IL-17–producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23–dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Fritz, J.H. et al. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity 26, 445–459 (2007).
van Beelen, A.J. et al. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity 27, 660–669 (2007).
Cho, J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466 (2008).
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003).
Gibson, D.L. et al. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium–induced colitis. Cell. Microbiol. 10, 618–631 (2008).
Acknowledgements
We thank L. Morikawa for help with preparing histological sections and K. Banks for help with animal experimentation. We also are very grateful to those individuals who volunteered for intestinal biopsies. Nod2−/− mice were provided by J.P. Hugot at Institut National de la Santé et de la Recherche Médicale U843. C. rodentium strain DBS100 was provided by B. Finlay at the University of British Columbia. This work was supported by a grant from the Crohn's and Colitis Foundation of Canada to S.E.G., a Crohn's and Colitis Foundation of Canada and a Canadian Institute of Health Research operating grant (480142) to D.J.P. and by a Canadian Institutes of Health Research grant (HET-85518) and an Ontario HIV Treatment Network grant (OGB-G123) to R.K. K.G. was supported by a Canadian Association of Gastroenterology/Canadian Institutes of Health Research postdoctoral research award, and S.J.R. was supported by a Fonds de la Recherche en Santé du Québec graduate scholarship.
Author information
Authors and Affiliations
Contributions
K.G. and S.J.R. designed and performed all experiments and wrote the manuscript. J.G.M. designed and performed mouse experiments. C.S. performed pathological scoring analysis. L.L.B. generated the Nod1−/−Nod2−/− mice. J.H.C. and S.J.R. performed microbiota analysis. C.J.K. and R.K. provided human colonic samples. D.J.P. and S.E.G. directed the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Methods (PDF 2371 kb)
Rights and permissions
About this article
Cite this article
Geddes, K., Rubino, S., Magalhaes, J. et al. Identification of an innate T helper type 17 response to intestinal bacterial pathogens. Nat Med 17, 837–844 (2011). https://doi.org/10.1038/nm.2391
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2391
This article is cited by
-
NOD2 signaling in CD11c + cells is critical for humoral immune responses during oral vaccination and maintaining the gut microbiome
Scientific Reports (2022)
-
Age-associated impairment of T cell immunity is linked to sex-dimorphic elevation of N-glycan branching
Nature Aging (2022)
-
Citrobacter rodentium–host–microbiota interactions: immunity, bioenergetics and metabolism
Nature Reviews Microbiology (2019)
-
Diverse Activity of IL-17+ Cells in Chronic Skin and Mucosa Graft-Versus-Host Disease
Archivum Immunologiae et Therapiae Experimentalis (2019)
-
Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities
Gut Pathogens (2018)