Abstract
To identify new genetic risk factors for Vogt-Koyanagi-Harada (VKH) syndrome, we conducted a genome-wide association study of 2,208,258 SNPs in 774 cases and 2,009 controls with follow-up in a collection of 415 cases and 2,006 controls and a further collection of 349 cases and 1,588 controls from a Han Chinese population. We identified three loci associated with VKH syndrome susceptibility (IL23R-C1orf141, rs117633859, Pcombined = 3.42 × 10−21, odds ratio (OR) = 1.82; ADO-ZNF365-EGR2, rs442309, Pcombined = 2.97 × 10−11, OR = 1.37; and HLA-DRB1/DQA1, rs3021304, Pcombined = 1.26 × 10−118, OR = 2.97). The five non-HLA genes were all expressed in human iris tissue. IL23R was also expressed in the ciliary body, and EGR2 was expressed in the ciliary body and choroid. The risk G allele of rs117633859 in the promoter region of IL23R exhibited low transcriptional activation in a cell-based reporter assay and was associated with diminished IL23R mRNA expression in human peripheral blood mononuclear cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Yang, P. et al. Clinical characteristics of Vogt-Koyanagi-Harada syndrome in Chinese patients. Ophthalmology 114, 606–614 (2007).
Moorthy, R.S., Inomata, H. & Rao, N.A. Vogt-Koyanagi-Harada syndrome. Surv. Ophthalmol. 39, 265–292 (1995).
Kim, M.H. et al. Association of HLA with Vogt-Koyanagi-Harada syndrome in Koreans. Am. J. Ophthalmol. 129, 173–177 (2000).
Weisz, J.M. et al. Association between Vogt-Koyanagi-Harada syndrome and HLA-DR1 and -DR4 in Hispanic patients living in southern California. Ophthalmology 102, 1012–1015 (1995).
Zhao, M., Jiang, Y. & Abrahams, I.W. Association of HLA antigens with Vogt-Koyanagi-Harada syndrome in a Han Chinese population. Arch. Ophthalmol. 109, 368–370 (1991).
Zhang, X.Y., Wang, X.M. & Hu, T.S. Profiling human leukocyte antigens in Vogt-Koyanagi-Harada syndrome. Am. J. Ophthalmol. 113, 567–572 (1992).
Islam, S.M. et al. HLA class II genes in Vogt-Koyanagi-Harada disease. Invest. Ophthalmol. Vis. Sci. 35, 3890–3896 (1994).
Hou, S. et al. Small ubiquitin-like modifier 4 (SUMO4) polymorphisms and Vogt-Koyanagi-Harada (VKH) syndrome in the Chinese Han population. Mol. Vis. 14, 2597–2603 (2008).
Chu, M. et al. Elevated serum osteopontin levels and genetic polymorphisms of osteopontin are associated with Vogt-Koyanagi-Harada disease. Invest. Ophthalmol. Vis. Sci. 52, 7084–7089 (2011).
Du, L. et al. Association of the CTLA-4 gene with Vogt-Koyanagi-Harada syndrome. Clin. Immunol. 127, 43–48 (2008).
Zhang, C. et al. MIF gene polymorphisms confer susceptibility to Vogt-Koyanagi-Harada syndrome in a Han Chinese population. Invest. Ophthalmol. Vis. Sci. 54, 7734–7738 (2013).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Wang, K., Li, M. & Bucan, M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 81, 1278–1283 (2007).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Murakami, S., Inaba, Y., Mochizuki, M., Nakajima, A. & Urayama, A. A nationwide survey on the occurrence of Vogt-Koyanagi-Harada disease in Japan. Jpn. J. Ophthalmol. 38, 208–213 (1994).
McMenamin, P.G., Crewe, J., Morrison, S. & Holt, P.G. Immunomorphologic studies of macrophages and MHC class II–positive dendritic cells in the iris and ciliary body of the rat, mouse, and human eye. Invest. Ophthalmol. Vis. Sci. 35, 3234–3250 (1994).
Wasserman, W.W. & Sandelin, A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5, 276–287 (2004).
Stranger, B.E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (2007).
Stranger, B.E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012).
Jiang, Z., Yang, P., Hou, S., Li, F. & Zhou, H. Polymorphisms of IL23R and Vogt-Koyanagi-Harada syndrome in a Chinese Han population. Hum. Immunol. 71, 414–417 (2010).
Mizuki, N. et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet's disease susceptibility loci. Nat. Genet. 42, 703–706 (2010).
Remmers, E.F. et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet's disease. Nat. Genet. 42, 698–702 (2010).
Jiang, Z. et al. IL23R gene confers susceptibility to Behcet's disease in a Chinese Han population. Ann. Rheum. Dis. 69, 1325–1328 (2010).
Xavier, J.M. et al. Association study of IL10 and IL23R-IL12RB2 in Iranian patients with Behcet's disease. Arthritis Rheum. 64, 2761–2772 (2012).
Kirino, Y. et al. Targeted resequencing implicates the familial Mediterranean fever gene MEFV and the toll-like receptor 4 gene TLR4 in Behcet disease. Proc. Natl. Acad. Sci. USA 110, 8134–8139 (2013).
Hughes, T. et al. Identification of multiple independent susceptibility loci in the HLA region in Behcet's disease. Nat. Genet. 45, 319–324 (2013).
Kirino, Y. et al. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 (2013).
Duerr, R.H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
McGovern, D.P. et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 (2010).
Silverberg, M.S. et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 41, 216–220 (2009).
Burton, P.R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Rioux, J.D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Evans, D.M. et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 (2011).
Nair, R.P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Haritunians, T. et al. Variants in ZNF365 isoform D are associated with Crohn's disease. Gut 60, 1060–1067 (2011).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Hirota, T. et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 44, 1222–1226 (2012).
Sun, L.D. et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat. Genet. 43, 690–694 (2011).
Lindström, S. et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat. Genet. 43, 185–187 (2011).
Okamura, T., Fujio, K., Sumitomo, S. & Yamamoto, K. Roles of LAG3 and EGR2 in regulatory T cells. Ann. Rheum. Dis. 71 (suppl. 2), i96–i100 (2012).
Li, S. et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 37, 685–696 (2012).
Read, R.W. et al. Revised diagnostic criteria for Vogt-Koyanagi-Harada disease: report of an international committee on nomenclature. Am. J. Ophthalmol. 131, 647–652 (2001).
International Study Group for Behçet's Disease. Criteria for diagnosis of Behcet's disease. Lancet 335, 1078–1080 (1990).
Howie, B., Marchini, J. & Stephens, M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470 (2011).
Zheng, X. et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 14, 192–200 (2014).
Acknowledgements
This work was supported by the Natural Science Foundation Major International (Regional) Joint Research Project (81320108009) (P.Y.), the National Basic Research Program of China (973 Program) (2011CB510200) (P.Y.), the Key Project of the Natural Science Foundation (81130019) (P.Y.), the National Natural Science Foundation Project (31370893 (P.Y.), 81270990 (S.H.) and 81025006 (Z. Yang)), the Research Fund for the Doctoral Program of Higher Education of China (20115503110002) (P.Y.), the Clinic Key Project of the Ministry of Health (201002019) (P.Y.), the Basic Research program of Chongqing (cstc2013jcyjC10001) (P.Y.), the Chongqing Key Laboratory of Ophthalmology (CSTC, 2008CA5003) (P.Y.), the National Key Clinical Specialties Construction Program of China (P.Y.), the Key Project of Health Bureau of Chongqing (2012-1-003) (P.Y.), the Fund for PAR-EU Scholars Program (P.Y.) and The Youth Outstanding-notch Talent Support Program of Chongqing (S.H.). We thank H. Zhou (Zhongshan Ophthalmic Center, Sun Yat-sen University), X. Liu (Department of Ophthalmology, The Second Hospital of Jilin University), Y. Wang (The Eye Hospital of Wenzhou Medical University), H. Wang (Department of Ophthalmology, Beijing Tongren Hospital, Capital Medical University), L. Xing (Ophthalmology of Department, the First Affiliated Hospital, Harbin Medical University), R. Zhang (Department of Ophthalmology, Eye and ENT Hospital of Fudan University), Y. Shi (Department of Ophthalmology, Shanxi Eye Hospital) and C. Zhao (Department of Ophthalmology, Nanjing General Hospital of the Nanjing Military Region) for helping with sample collection, and we also thank the individuals, their families and friends who participated in this project. Some samples of cases and controls were collected in the Zhongshan Ophthalmic Center, Sun Yat-sen University. We thank Genergy Bio-technology Corporation (Shanghai) and CapitalBio Corporation (Beijing) for helping with the microarray experiment, and we also thank Beijing Genomics Institute for luciferase reporter analysis.
Author information
Authors and Affiliations
Contributions
P.Y., Z. Yang and S.H. conceived and designed the study. S.H., M.W. and J.X. analyzed the GWAS data. P.Y., Z. Yang, L.D., C.P.P., Meifen Zhang, W.Z., Minglian Zhang, Q. Zhang, K.H., Q. Zhou and H.L. recruited subjects and participated in the diagnostic evaluations. S.H., L.D., L.H., K.H., J.Q., C.W., Y.T., Z. Ye, Y.Z., Y.L., L.B. and D.L. contributed to the genotyping. L.L., H.Y. and Q.C. contributed to the reagents, materials and analysis tools. S.H., B.L., B.G. and D.L. contributed to the RT-PCR or real-time PCR. S.H. and P.Y. drafted the manuscript. B.L., C.P.P., J.X., A.K. and Z. Yang helped revise the manuscript. P.Y., Z. Yang and S.H. obtained funding for this study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 2 The principal component analysis (PCA) of GWAS samples and HapMap 270 individuals (JPT+CHB, CEU and YRI).
Distribution of the subjects used in GWAS and four HapMap polulations.
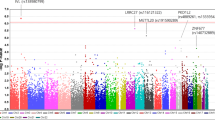
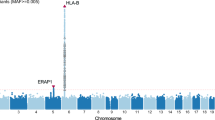
Supplementary Figure 4 Genome-wide association results for non-HLA SNPs.
(a) Q-Q plots of the observed P values (y axis) versus the expected P-values (x axis) from genome-wide association results for non-HLA SNPs. (b) Manhattan plot of P values on the –log10 scale for 2,199,482 non-HLA SNPs (removed SNPs at chr 6: 28-34M) in the GWAS stage (774 VKH cases and 2,009 normal controls). The red line represents P = 1.0 × 10−8, and the blue dashed line represents P = 1.0 × 10−6.
Supplementary Figure 5 Regional plot of association results for the HLA region.
Results (-log10 P) are shown for SNPs in the 6p21.3 region flanking 400 kb on either side of the index SNPs (purple diamond). The association results for both genotyped and imputed SNPs are shown with the recombination rates estimated from the 1000 Genomes Project CHB and JPT data. The index SNP is shown in diamonds, and the r2 values of the remaining SNPs are indicated by color. The genes within the region are annotated and indicated by arrows.
Supplementary Figure 6 LD block at the IL23R-C1orf141 locus flanking SNP rs117633859 in the Han Chinese population.
Haplotype LD block was estimated for the SNPs at the IL23R-C1orf141 locus using the 1000 Genomes Project CHB data by Haploview software. The r2 values are shown in blocks. D’ ≥ 0.80, red; 0.5 ≤ D’ < 0.8, pink; 0.2 ≤ D’ < 0.5, blue; D’ < 0.2, white.
Supplementary Figure 7 The expression of IL23R in human uveal tissues and the association of SNP rs117633859 near IL23R with mRNA expression levels in normal control peripheral blood mononuclear cells (PBMCs) and luciferase activity analysis.
(a,b) The mRNA expression of IL23R in the human uveal tissues including the iris, ciliary body and choroid from four controls (lanes 1-4) using RT-PCR. (c) The mRNA expression of IL23R according to rs117633859 genotypes in human PBMCs from normal controls. A Mann-Whitney nonparametric test was used to test for significance among different genotypes of rs117633859. P < 0.05 was considered significant. Error bar: s.d. (d) The ~5 kb sequences of IL23R carrying the risk G allele or the A allele of SNP rs117633859 were cloned into a pGL3-basic vector. Luciferase activity was determined using a Modulus system (Turner Biosystems). The experiment was repeated twelve times (n = 12). An independent-samples T test was used to test for the significance of luciferase activity between risk and protective alleles of rs117633859. Error bar: s.d.
Supplementary Figure 8 The expression of C1orf141, ADO, ZNF365, EGR2 and IL23R in the human uveal tissues.
(a–e) The mRNA expression of C1orf141 (a), ADO (b), ZNF365 (c), EGR2 (d) and β-actin (e) in uveal tissues (iris, ciliary body and choroid) from four controls (lanes 1-4). (f) The mRNA expression of IL23R in the iris between VKH patients without active inflammation and controls. The iris tissue was obtained from fourteen eye donors without ocular inflammatory disease or a history of eye disease and seven VKH patients without active inflammation (obtained during iridectomy surgery to prevent secondary glaucoma). The experiment was repeated triple times (n = 3). An independent-samples T test was used to test for the significance of IL23R between VKH patients and controls.
Supplementary Figure 9 A locus-zoom plot of the IL23R expression quantitative trait locus (eQTL) effect around SNP rs117633859.
(a) The locus-zoom plot of the IL23R eQTL effect using HapMap Han Chinese in Beijing, China (CHB) from GEO data sets (GSE6536). (b) The locus-zoom plot of the IL23R eQTL effect using HapMap Han Chinese in Beijing, China (CHB) from HAPMAP3_EXPRESSION database (E-MTAB-264). Twenty SNPs including rs77258390, rs3762318, rs12563505, rs34017352, rs114661385, rs117301158, rs115009271, rs76091599, rs78865162, rs78917656, rs78597810, rs12568393, rs12561798, rs76436269, rs6693659, rs78377598, rs12564219, rs12566159, rs117282985, rs117633859 (from left to right) were enrolled in the eQTLs analysis.
Supplementary Figure 10 LD block of SNPs at the IL23R locus associated with multiple autoimmune diseases.
(a) Haplotype LD block was estimated for the IL23R SNPs associated with VKH syndrome in the present study and reported by previous study (Jiang et al. Hum. Immunol. 71, 414-417, 2010) using the 1000 Genomes Project CHB data by Haploview software. The r2 values are shown in blocks. D’ ≥ 0.80, red; 0.5 ≤ D’ < 0.8, pink; 0.2 ≤ D’ < 0.5, blue; D’ < 0.2, white. (b,c) Haplotype LD block was estimated for the IL23R SNPs associated with VKH syndrome and other autoimmune disease including ankylosing spondylitis, Crohn’s disease, ulcerative colitis and psoriasis using the present GWAS controls data (b) and cases data (c) by Haploview software. The r2 values are shown in blocks. D’ ≥ 0.80, red; 0.5 ≤ D’ < 0.8, pink; 0.2 ≤ D’ < 0.5, blue; D’ < 0.2, white. SNP rs11209026 associated with a variety of autoimmune diseases such as Crohn’s disease, psoriasis and ankylosing spondylitis is monomorphic in the Han Chinese population.
Supplementary Figure 11 Association of SNP rs442309 in ADO-ZNF365-EGR2 with gene expression levels in human lymphoblastoid cell lines.
The mRNA expression of ADO, ZNF365 and EGR2 according to rs442309 genotypes using HapMap CHB and JPT (downloaded from HAPMAP3_EXPRESSION database (E-MTAB-264)).
Supplementary Figure 12 Cluster plot of 384 DNA samples for SNP rs117633859 by TaqMan assay using AB7900 Real-Time PCR system.
Green color stand for homozygote AA. Red color stands for heterozygote AG. Blue color stands for homozygote GG. Gray color stands for no call.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1-12, Supplementary Tables 1-3, 5, 6, 9-11, 14 and 15 and Supplementary Note (PDF 2903 kb)
Supplementary Table 4
Results for associated SNPs at 1p31.2 and 10q21.3 in GWAS with VKH syndrome (P < 0.0001) (XLSX 17 kb)
Supplementary Table 7
Results for three associated loci after adjusting for two array platforms (XLSX 18 kb)
Supplementary Table 8
Summary of associated SNPs in the HLA region with VKH syndrome (XLSX 28 kb)
Supplementary Table 12
Summary of association results for IL23R and ADO-EGR2-ZNF365 genes with human diseases (XLSX 26 kb)
Supplementary Table 13
Summary of association results for multiple autoimmune diseases by GWAS (XLSX 53 kb)
Rights and permissions
About this article
Cite this article
Hou, S., Du, L., Lei, B. et al. Genome-wide association analysis of Vogt-Koyanagi-Harada syndrome identifies two new susceptibility loci at 1p31.2 and 10q21.3. Nat Genet 46, 1007–1011 (2014). https://doi.org/10.1038/ng.3061
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3061
This article is cited by
-
Kernel-based genetic association analysis for microbiome phenotypes identifies host genetic drivers of beta-diversity
Microbiome (2023)
-
Genetic association of PRKCD and CARD9 polymorphisms with Vogt–Koyanagi–Harada disease in the Chinese Han population
Human Genomics (2023)
-
A randomized non-inferiority trial of therapeutic strategy with immunosuppressants versus biologics for Vogt-Koyanagi-Harada disease
Nature Communications (2023)
-
A de novo missense mutation in MPP2 confers an increased risk of Vogt–Koyanagi–Harada disease as shown by trio-based whole-exome sequencing
Cellular & Molecular Immunology (2023)
-
Progress in the genetics of uveitis
Genes & Immunity (2022)