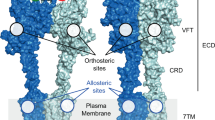
Abstract
Glutamate receptors are the predominant mediators of excitatory synaptic signals in the central nervous system and are important in learning and memory as well as in diverse neuropathologies including epilepsy and ischemia1,2,3,4,5. Their primary function is to receive the chemical signal glutamate (1), which binds to an extracellular domain in the receptor, and convert it into an electrical signal through the formation of cation-permeable transmembrane channels6,7,8,9. Recently described end-state apo and ligated structures of the ligand-binding domain of a rat glutamate receptor provide a first view of specific molecular interactions between the ligand and the receptor that are central to the allosteric regulation of function in this protein7,10,11,12,13,14,15. Yet there is little information on the mechanism and the structures of intermediates (if any) formed during the ligand-binding process16. Here we have used time-resolved vibrational spectroscopy to show that the process involves a sequence of interleaved ligand and protein changes that starts with the docking of glutamate at the α-carboxylate moiety and ends with the establishment of the interactions between the γ-carboxylate of glutamate and the protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hollmann, M. & Heinemann, S. Cloned glutamate receptors. Annu. Rev. Neurosci. 17, 31–108 (1994).
Nakanishi, S. & Masu, M. Molecular diversity glutamate receptors and their physiological functions. Annu. Rev. Biophys. Biomol. Struct. 23, 329–348 (1994).
Dingledine, R., Borges, K., Bowie, D. & Traynelis, S.F. The glutamate receptor ion channels. Pharmacol. Rev. 51, 7–61 (1999).
Riedel, G., Platt, B. & Micheau, J. Glutamate receptor function in learning and memory. Behav. Brain Res. 140, 1–47 (2003).
Bergink, V., van Megen, H.J. & Westenberg, H.G. Glutamate and anxiety. Eur. Neuropsychopharmacol. 14, 175–183 (2004).
Madden, D.R. The structure and function of glutamate receptor ion channels. Nat. Rev. Neurosci. 3, 91–101 (2002).
McFeeters, R.L. & Oswald, R.E. Emerging structural explanations of ionotropic glutamate receptor function. FASEB J. 18, 428–438 (2004).
Mayer, M.L. & Armstrong, N. Structure and function of glutamate receptor ion channels. Annu. Rev. Physiol. 66, 161–181 (2004).
Mayer, M.L. Glutamate receptor ion channels. Curr. Opin. Neurobiol. 15, 282–288 (2005).
Armstrong, N. & Gouaux, E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000).
Jin, R., Banke, T.G., Mayer, M.L., Traynelis, S.F. & Gouaux, E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6, 803–810 (2003).
Sun, Y. et al. Mechanism of glutamate receptor desensitization. Nature 417, 245–253 (2002).
Cheng, Q. & Jayaraman, V. Chemistry and conformation of the ligand-binding domain of GluR2 subtype of glutamate receptors. J. Biol. Chem. 279, 26346–26350 (2004).
Jayaraman, V. Spectroscopic and kinetic methods for ligand-protein interactions of glutamate receptor. Methods Enzymol. 380, 170–187 (2004).
Du, M., Reid, S.A. & Jayaraman, V. Conformational changes in the ligand-binding domain of a functional ionotropic glutamate receptor. J. Biol. Chem. 280, 8633–8636 (2005).
Abele, R., Keinänen, K. & Madden, D.R. Agonist-induced isomerization in a glutamate receptor ligand-binding domain: a kinetic and mutagenetic analysis. J. Biol. Chem. 275, 21355–21363 (2000).
Madden, D.R., Abele, R., Andersson, A. & Keinänen, K. Large-scale expression and thermodynamic characterization of a glutamate receptor agonist-binding domain. Eur. J. Biochem. 267, 4281–4289 (2000).
Dunn, B.C. & Eyring, E.M. Stopped-flow rapid-scan Fourier transform infrared spectroscopy. Appl. Spectrosc. 53, 292–296 (1999).
Armstrong, N., Mayer, M. & Gouaux, E. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc. Natl. Acad. Sci. USA 100, 5736–5741 (2003).
Wieboldt, R. et al. Photolabile precursors of glutamate: Synthesis, photochemical properties, and activation of glutamate receptors on a microsecond time scale. Proc. Natl. Acad. Sci. USA 91, 8752–8756 (1994).
Cheng, Q., Steinmetz, M.G. & Jayaraman, V. Photolysis of γ-(α-carboxy-2-nitrobenzyl)-l-glutamic acid investigated in the microsecond time scale by time-resolved FTIR. J. Am. Chem. Soc. 124, 7676–7677 (2002).
Acknowledgements
This work was supported by the National Science Foundation (MCB-0444352), the National Institutes of Health (1R03AA015682-01) and the American Heart Foundation, Texas Affiliate.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Time-resolved FTIR difference spectra showing the evolution of the glutamate and protein vibrational modes during agonist binding to the GluR2S1S2. (PDF 19 kb)
Supplementary Fig. 2
Difference FTIR spectrum between glutamate-bound and unligated form of GluR2 S1S2-E705D protein in D2O, and glutamate-bound and unligated form of GluR2 S1S2-E705D protein in H2O. (PDF 11 kb)
Supplementary Fig. 3
Difference FTIR spectrum between glutamate and D2O buffer, glutamate and H2O buffer, [γ-13C]glutamate and H2O buffer and [α-13C]glutamate and H2O buffer (PDF 23 kb)
Rights and permissions
About this article
Cite this article
Cheng, Q., Du, M., Ramanoudjame, G. et al. Evolution of glutamate interactions during binding to a glutamate receptor. Nat Chem Biol 1, 329–332 (2005). https://doi.org/10.1038/nchembio738
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio738
This article is cited by
-
cAMP binding to closed pacemaker ion channels is non-cooperative
Nature (2021)
-
Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels
Nature Structural & Molecular Biology (2016)
-
The hidden energetics of ligand binding and activation in a glutamate receptor
Nature Structural & Molecular Biology (2011)
-
Structural landscape of isolated agonist-binding domains from single AMPA receptors
Nature Chemical Biology (2011)
-
New light on an open-and-shut case
Nature Chemical Biology (2005)