Abstract
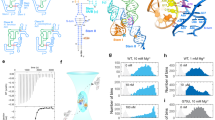
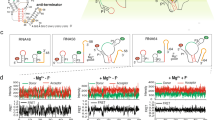
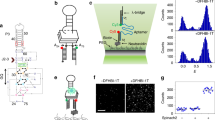
Twister is a small ribozyme present in almost all kingdoms of life that rapidly self-cleaves in variety of divalent metal ions. We used activity assays, bulk FRET and single-molecule FRET (smFRET) to understand how different metal ions promote folding and self-cleavage of the Oryza sativa twister ribozyme. Although most ribozymes require additional Mg2+ for catalysis, twister inverts this expectation, requiring 20–30 times less Mg2+ to self-cleave than to fold. Transition metals such as Co2+, Ni2+ and Zn2+ activate twister more efficiently than Mg2+ ions. Although twister is fully active in ≤ 0.5 mM MgCl2, smFRET experiments showed that the ribozyme visits the folded state infrequently under these conditions. Comparison of folding and self-cleavage rates indicates that most folding events lead to catalysis, which correlates with metal bond strength. Thus, the robust activity of twister reports on transient metal ion binding under physiological conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Jimenez, R.M., Polanco, J.A. & Lupták, A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 40, 648–661 (2015).
Roth, A. et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 10, 56–60 (2014).
Weinberg, Z. et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 11, 606–610 (2015).
Furukawa, K. et al. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol. Cell 57, 1088–1098 (2015).
Price, I.R., Gaballa, A., Ding, F., Helmann, J.D. & Ke, A. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell 57, 1110–1123 (2015).
DeRose, V.J. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 13, 317–324 (2003).
Liu, Y., Wilson, T.J., McPhee, S.A. & Lilley, D.M. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 10, 739–744 (2014).
Ren, A. et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 5, 5534 (2014).
Wilson, T.J., Liu, Y., Domnick, C., Kath-Schorr, S. & Lilley, D.M. The novel chemical mechanism of the twister ribozyme. J. Am. Chem. Soc. 138, 6151–6162 (2016).
Gaines, C.S. & York, D.M. Ribozyme catalysis with a twist: active state of the twister ribozyme in solution predicted from molecular simulation. J. Am. Chem. Soc. 138, 3058–3065 (2016).
Zhang, S. et al. Role of the active site guanine in the glmS ribozyme self-cleavage mechanism: quantum mechanical/molecular mechanical free energy simulations. J. Am. Chem. Soc. 137, 784–798 (2015).
Gebetsberger, J. & Micura, R. Unwinding the twister ribozyme: from structure to mechanism. Wiley Interdiscip. Rev. RNA 8 (2017).
Vušurović, N., Altman, R.B., Terry, D.S., Micura, R. & Blanchard, S.C. Pseudoknot formation seeds the twister ribozyme cleavage reaction coordinate. J. Am. Chem. Soc. 139, 8186–8193 (2017).
Košutić, M. et al. A mini-twister variant and impact of residues/cations on the phosphodiester cleavage of this ribozyme class. Angew. Chem. Int. Edn. Engl. 54, 15128–15133 (2015).
Ucisik, M.N., Bevilacqua, P.C. & Hammes-Schiffer, S. Molecular dynamics study of twister ribozyme: role of Mg2+ ions and the hydrogen-bonding network in the active site. Biochemistry 55, 3834–3846 (2016).
Alatossava, T., Jütte, H., Kuhn, A. & Kellenberger, E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162, 413–419 (1985).
Froschauer, E.M., Kolisek, M., Dieterich, F., Schweigel, M. & Schweyen, R.J. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol. Lett. 237, 49–55 (2004).
Zhong, W., Schobert, C. & Komor, E. Transport of magnesium ions in the phloem of Ricinus communis L. seedlings. Planta 190, 114–119 (1993).
Ha, T. et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 93, 6264–6268 (1996).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Kilburn, D., Roh, J.H., Guo, L., Briber, R.M. & Woodson, S.A. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 132, 8690–8696 (2010).
Paudel, B.P. & Rueda, D. Molecular crowding accelerates ribozyme docking and catalysis. J. Am. Chem. Soc. 136, 16700–16703 (2014).
Dupuis, N.F., Holmstrom, E.D. & Nesbitt, D.J. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. USA 111, 8464–8469 (2014).
Bokinsky, G. et al. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. USA 100, 9302–9307 (2003).
Leffler, J.E. Parameters for the description of transition states. Science 117, 340–341 (1953).
Tanford, C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv. Protein Chem. 24, 1–95 (1970).
Jackson, S.E. & Fersht, A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 30, 10428–10435 (1991).
Koculi, E., Thirumalai, D. & Woodson, S.A. Counterion charge density determines the position and plasticity of RNA folding transition states. J. Mol. Biol. 359, 446–454 (2006).
Sosnick, T.R. Kinetic barriers and the role of topology in protein and RNA folding. Protein Sci. 17, 1308–1318 (2008).
Irving, H. & Willams, R.J.P. Order of stability of metal complexes. Nature 162, 746–747 (1948).
Woodson, S.A. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9, 104–109 (2005).
Draper, D.E., Grilley, D. & Soto, A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34, 221–243 (2005).
Ward, W.L., Plakos, K. & DeRose, V.J. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem. Rev. 114, 4318–4342 (2014).
Eiler, D., Wang, J. & Steitz, T.A. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc. Natl. Acad. Sci. USA 111, 13028–13033 (2014).
Kobori, S. & Yokobayashi, Y. High-throughput mutational analysis of a twister ribozyme. Angew. Chem. Int. Edn Engl. 55, 10354–10357 (2016).
Saunders, A.M. & DeRose, V.J. Beyond Mg2+: functional interactions between RNA and transition metals. Curr. Opin. Chem. Biol. 34, 152–158 (2016).
Dahm, S.C. & Uhlenbeck, O.C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30, 9464–9469 (1991).
Pan, T. & Uhlenbeck, O.C. A small metalloribozyme with a two-step mechanism. Nature 358, 560–563 (1992).
Leonarski, F., D'Ascenzo, L. & Auffinger, P. Binding of metals to purine N7 nitrogen atoms and implications for nucleic acids: A CSD survey. Inorganica Chim. Acta 452, 82–89 (2016).
Sigel, R.K. & Sigel, H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 43, 974–984 (2010).
Kobayashi, N.I. & Tanoi, K. Critical issues in the study of magnesium transport systems and magnesium deficiency symptoms in plants. Int. J. Mol. Sci. 16, 23076–23093 (2015).
Yoneyama, T., Ishikawa, S. & Fujimaki, S. Route and regulation of zinc, cadmium, and iron transport in rice plants (Oryza sativa L.) during vegetative growth and grain filling: metal transporters, metal speciation, grain Cd reduction and Zn and Fe biofortification. Int. J. Mol. Sci. 16, 19111–19129 (2015).
Takahashi, R., Bashir, K., Ishimaru, Y., Nishizawa, N.K. & Nakanishi, H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 7, 1605–1607 (2012).
Solomatin, S. & Herschlag, D. Methods of site-specific labeling of RNA with fluorescent dyes. Methods Enzymol. 469, 47–68 (2009).
Hua, B. et al. An improved surface passivation method for single-molecule studies. Nat. Methods 11, 1233–1236 (2014).
Kim, H. et al. Protein-guided RNA dynamics during early ribosome assembly. Nature 506, 334–338 (2014).
McKinney, S.A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Acknowledgements
The authors thank K. Sarkar and S. Abeysirigunawardena for their assistance and M. Greenberg, K. Karlin and J. Morrow for helpful discussion. This work was supported by a grant from the National Science Foundation (MCB-1616081 to S.W.) and the US National Institutes of Health (GM 065367 to T.H.).
Author information
Authors and Affiliations
Contributions
S.P., T.H. and S.A.W. designed the experiments; S.P. and B.H. performed the single-molecule experiments and analyzed the data; S.P. and D.Z. performed activity assays; S.P. performed other experiments; S.P., T.H. and S.A.W. wrote the manuscript; and all of the authors interpreted the data and reviewed the text and figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–11. (PDF 1396 kb)
Reporting Summary
Reporting Summary. (PDF 133 kb)
Rights and permissions
About this article
Cite this article
Panja, S., Hua, B., Zegarra, D. et al. Metals induce transient folding and activation of the twister ribozyme. Nat Chem Biol 13, 1109–1114 (2017). https://doi.org/10.1038/nchembio.2459
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2459
This article is cited by
-
RNA compaction and iterative scanning for small RNA targets by the Hfq chaperone
Nature Communications (2024)
-
Metal ions and sugar puckering balance single-molecule kinetic heterogeneity in RNA and DNA tertiary contacts
Nature Communications (2020)