Abstract
The phytohormone auxin is a key developmental signal in plants. So far, only auxin perception has been described to trigger the release of transcription factors termed AUXIN RESPONSE FACTORs (ARFs) from their AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressor proteins. Here, we show that phosphorylation of ARF7 and ARF19 by BRASSINOSTEROID-INSENSITIVE2 (BIN2) can also potentiate auxin signalling output during lateral root organogenesis. BIN2-mediated phosphorylation of ARF7 and ARF19 suppresses their interaction with AUX/IAAs, and subsequently enhances the transcriptional activity to their target genes LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16) and LBD29. In this context, BIN2 is under the control of the TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)–TDIF RECEPTOR (TDR) module. TDIF-initiated TDR signalling directly acts on BIN2-mediated ARF phosphorylation, leading to the regulation of auxin signalling during lateral root development. In summary, this study delineates a TDIF–TDR–BIN2 signalling cascade that controls regulation of ARF and AUX/IAA interaction independent of auxin perception during lateral root development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Vanneste, S. & Friml, J. Auxin: a trigger for change in plant development. Cell 136, 1005–1016 (2009).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005).
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005).
Lau, S., Jurgens, G. & De Smet, I. The evolving complexity of the auxin pathway. Plant Cell 20, 1738–1746 (2008).
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008).
Villalobos, L. I. A. C. et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 (2012).
De Rybel, B. et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706 (2010).
De Smet, I. Lateral root initiation: one step at a time. New Phytol. 193, 867–873 (2012).
De Smet, I. et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 2705–2710 (2010).
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A. & Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130 (2007).
Tatematsu, K. et al. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393 (2004).
Clevers, H. Wnt/ β-catenin signaling in development and disease. Cell 127, 469–480 (2006).
Dornelas, M. C., Wittich, P., von Recklinghausen, I., van Lammeren, A. & Kreis, M. Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family. Plant Mol. Biol. 39, 137–147 (1999).
Saidi, Y., Hearn, T. J. & Coates, J. C. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 17, 39–46 (2012).
Kim, T. W., Michniewicz, M., Bergmann, D. C. & Wang, Z. Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422 (2012).
Li, J. M. & Nam, K. H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301 (2002).
Gudesblat, G. E. et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554 (2012).
Ryu, H. et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762 (2007).
Wang, Z. Y. et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513 (2002).
Yin, Y. et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191 (2002).
Kim, T. W. et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260 (2009).
Perez-Perez, J. M., Ponce, M. R. & Micol, J. L. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242, 161–173 (2002).
Vert, G., Walcher, C. L., Chory, J. & Nemhauser, J. L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl Acad. Sci. USA 105, 9829–9834 (2008).
Choe, S. et al. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 130, 1506–1515 (2002).
Li, J., Nam, K. H., Vafeados, D. & Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22 (2001).
Yan, Z., Zhao, J., Peng, P., Chihara, R. K. & Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721 (2009).
De Rybel, B. et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604 (2009).
Choi, S. et al. BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J. 73, 380–391 (2012).
Nam, K. H. & Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 (2002).
Ryu, H., Kim, K., Cho, H. & Hwang, I. Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol. Cells 29, 291–296 (2010).
Okushima, Y. et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463 (2005).
Peret, B. et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408 (2009).
Hwang, I. & Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389 (2001).
Ruegger, M. et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207 (1998).
Hirakawa, Y. et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl Acad. Sci. USA 105, 15208–15213 (2008).
Mouchel, C. F., Osmont, K. S. & Hardtke, C. S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 (2006).
Bao, F. et al. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631 (2004).
Murphy, E., Smith, S. & De Smet, I. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24, 3198–3217 (2012).
Yaginuma, H., Hirakawa, Y., Kondo, Y., Ohashi-Ito, K. & Fukuda, H. A novel function of TDIF-related peptides: promotion of axillary bud formation. Plant Cell Physiol. 52, 1354–1364 (2011).
Goda, H. et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. The Plant J. 55, 526–542 (2008).
De Smet, I. et al. An easy and versatile embedding method for transverse sections. J. Microsc. 213, 76–80 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Chang, C. S., Maloof, J. N. & Wu, S. H. COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156, 228–239 (2011).
Acknowledgements
The authors thank D. Weijers for critical reading of the manuscript and useful suggestions. This work was supported by grants from the Advanced Biomass R&D Center (2010-0029720) and the National Research Foundation (20110000212) funded by the Korean Ministry of Education, Science and Technology. H.R. was supported by grants from the Next-Generation BioGreen 21 Program (PJ009516). H.C. and S.H. were recipients of a Brain Korea 21 fellowship. I.D.S. was supported by a BBSRC David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration Grant (PERG06-GA-2009-256354).
Author information
Authors and Affiliations
Contributions
H.C., H.R., S.H., J.P., K.H. and S.S. performed experiments. H.C., H.R., I.D.S. and I.H. designed experiments and analysed the data. D.A. analysed gene expression. S.R. and D.H. performed LC–MS/MS analysis. H.C., H.R., T.B., M.J.B., I.D.S. and I.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 The expression pattern of BIN2, ARF7 and ARF19 in root.
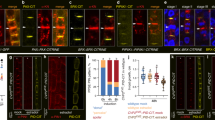
(a) BIN2 expression in basal meristem and LRP at different stages of lateral root development. Note that the expression is restricted to the basal part of LRP at stages IV-VII. Scale bar, 50 μm. (b,c) ARF7 (b) and ARF19 (c) expression in LRP at early stages of lateral root development. GUS activity driven by the ARF7 or ARF19 promoter was observed in early stages of LRP. Scale bar, 50 μm.
Supplementary Figure 2 BIN2 enhances auxin-response in plants.
(a) Three-day-old seedlings of pDR5-GUS and pDR5-GUS / bin2-1-gof grown on 10 μM NPA-containing medium were transferred to 1 μM NAA-containing medium, and further treated for 1, 3, and 5 h. The GUS activity of the seedlings was measured. Error bars indicate SEM (n = 3, 20 plants per n = 1, * for P<0.05 by student’s t-test). (b) The expression levels of LBD16 and LBD29 in 10-day-old bri1-116 and bin2-1-gof (+/+) mutants were determined by quantitative RT-PCR (n = 4, 10 plants per n = 1, * for P<0.05 by student’s t-tests). (c,d) The expression levels of LBD16 (left panel) and LBD29 (right panel) in 10-day-old WT and bin2-1-gof (c), dwf12-1D-gof (d), and bin2-3 bil1 bil2 mutants were determined by quantitative RT-PCR after auxin treatment for the indicated times. Error bars indicate SEM (n = 3, 10 plants per n = 1, * for P<0.05 by student’s t-tests). (e) Bikinin treatment represses LBD16 and LBD29 expression. Error bars indicate SEM (n = 3, 10 plants per n = 1, * for P<0.05 by student’s t-tests). (f) BIN2 enhances the activity of pGH3-LUC auxin-responsive reporter in a dose-dependent manner. Protoplasts were co-transfected with pGH3-LUC, indicated amount of BIN2-HA, and pUBQ10-GUS (internal control). Error bars indicate the SEM (n = 4, 20,000 cells per n = 1, * for P<0.05, ** for P<0.01 by student’s t-test). Statistics source data found in Supplementary Table 2. Details on the statistical analysis are found in Methods.
Supplementary Figure 3 BIN2 and BIL1 are involved in lateral root formation.
(a) BIN2 and BIL1 enhance auxin-responsive pGH3-LUC activity. Protoplasts were cotransfected with pUBQ10-GUS (internal control), pGH3-LUC and an effector plasmid harboring BIN2, BIL1 or BIL2. Error bars indicate SEM (n = 4, 20,000 cells per n = 1, * for P<0.05 by student’s t-test). (b) Isolation of bil1 knockout mutant. The transcripts of BIL1 in 7-day-old wild type (Ws-2) and bil1 (FLAG_426B01) were determined by semi-quantitative PCR. (c) Lateral root densities of twelve-day-old seedlings of WT (Ws-2), bin2-3-lof, bil1, bil1 bin2-3 and bin2-3 bil1 bil2 mutant. E, emerged lateral roots; NE, non-emerged LRP. Error bars indicate SEM (n = 15 plants, * for P<0.05, ** for P<0.01 by student’s t-test). (d) The transcripts of ARF7 in WT (Col-0), arf7-1 and arf7-1 bin2-1-gof were determined by semi-quantitative PCR. (e) The transcripts of ARF7 and ARF19 in WT (Col-0) and arf7-1 arf19-1 bin2-1-gof were determined by semi-quantitative PCR. Tubulin and Actin2 genes served as input controls. Details on the statistical analysis are found in Methods.
Supplementary Figure 4 BIN2 does not interact with AUX/IAA proteins.
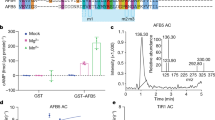
HA-tagged BIN2 was expressed in protoplasts. Protoplast lysates were incubated with GST-IAA1, GST-IAA3, or GST-IAA19 proteins for 1 h, and then precipitated with glutathione sepharose 4B. BIN2 proteins were detected with anti-HA antibody (upper and middle panels), and GST-tagged IAA1, IAA3, or IAA19 proteins were visualized by Coomassie blue staining (lower panel).
Supplementary Figure 5 BIN2-mediated phosphorylation of ARF7 attenuates its interaction with IAA1, IAA3, IAA14 and IAA18.
Protoplast lysates overexpressing ARF7-HA or ARF7-HA and BIN2-HA were incubated with purified GST-tagged IAA1, IAA3, IAA14, or IAA18 proteins and pulled down with glutathione sepharose 4B. The pulled-down proteins were analyzed with anti-HA antibody. The GST protein was used as a negative control. An asterisk indicates GST-IAAs proteins.
Supplementary Figure 6 (a) BIN2-mediated phosphorylation of ARF7 results in increased binding of IAA19 with TIR1.
Two more independent pull down experiments conducted in Fig. 6a are presented. HA-tagged ARF7 and TIR1 were co-transfected into protoplasts with or without BIN2-HA. After 6 h of incubation, protoplast lysates were incubated with purified GST-IAA19 proteins and 10 μM of MG132 for 1 h, and then pulled down with glutathione sepharose 4B. GST-IAA19-bound proteins were visualized with anti-HA antibody. GST protein was included as a negative control. Asterisks indicate GST-IAA19 proteins. The levels of GST-IAA19 pulled-down proteins were normalized to those of the input proteins. The normalized intensities of the pulled-down proteins in the absence of BIN2 were set to 1, and the relative protein intensities were presented under the corresponding protein bands. (b) Relative DII-VENUS intensities in seedlings of mock or 15 μMbikinin treated WT (Col-0). (n = 20 plants).
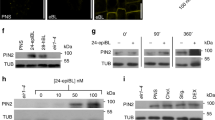
Supplementary Figure 7 Brassinosteroid is not involved in BIN2-mediated ARF7 activation.
(a,b) Brassinosteroid does not affect the phosphorylation status of ARF7. Seven-day-old transgenic seedlings harboring 35S-ARF7-HA (left panel), 35S-ARF7-HA / dwf12-1D-gof (b), or 35S-BES1-HA (a positive BR-responsive control, right panel) were incubated with or without 20 nM of epi-BL for 6 and 12 h. The phosphorylation status of HA-tagged ARF7 and BES1 was determined as the protein mobility shift with anti-HA antibody. (c) Bikinin inhibits BIN2-induced phosphorylation of ARF7. ARF7-HA was co-transfected with BIN2-HA into protoplasts and incubated with or without 30 μM of bikinin for 6 h. ARF7 proteins were visualized with anti-HA antibody. (d) Brassinosteroid could not suppress the BIN2 actions for enhancing the transcriptional activity of ARF7. ARF7-HA, pGH3-LUC and pUBQ10-GUS were cotransfected into protoplasts with or without BIN2-HA. The protoplasts were incubated with or without 1 μM epi-BL and 30 μM bikinin for 6 h. Error bars indicate SEM (n = 4, 20,000 cells per n = 1).
Supplementary information
Supplementary Information
Supplementary Information (PDF 1711 kb)
Supplementary Table 1
Supplementary Information (XLSX 41 kb)
Supplementary Table 2
Supplementary Information (XLSX 59 kb)
Rights and permissions
About this article
Cite this article
Cho, H., Ryu, H., Rho, S. et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16, 66–76 (2014). https://doi.org/10.1038/ncb2893
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2893
This article is cited by
-
Genome-wide identification and expression analysis of the Auxin-Response factor (ARF) gene family in Medicago sativa under abiotic stress
BMC Genomics (2023)
-
The phytohormones underlying the plant lateral root development in fluctuated soil environments
Plant and Soil (2023)
-
Effects of ectopic expression of WOX4 and WOX14 on stem cell maintenance and organogenesis of Arabidopsis thaliana
Acta Physiologiae Plantarum (2023)
-
A positive response of ginger root zone and rhizome development to suitable sowing depth
Protoplasma (2022)
-
Zea mays GSK2 gene is involved in brassinosteroid signaling
Plant Growth Regulation (2022)