Abstract
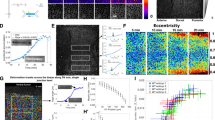
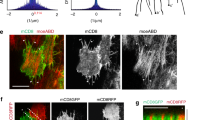
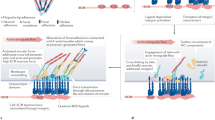
Regulated assembly and disassembly, or turnover, of integrin-mediated cell–extracellular matrix (ECM) adhesions is essential for dynamic cell movements and long-term tissue maintenance. For example, in Drosophila, misregulation of integrin turnover disrupts muscle–tendon attachment at myotendinous junctions (MTJs). We demonstrate that mechanical force, which modulates integrin activity, also regulates integrin and intracellular adhesion complex (IAC) turnover in vivo. Using conditional mutants to alter the tensile force on MTJs, we found that the proportion of IAC components undergoing turnover inversely correlated with the force applied on MTJs. This effect was disrupted by point mutations in β-integrin that interfere with ECM-induced conformational changes and activation of β-integrin or integrin-mediated cytoplasmic signalling. These mutants also disrupted integrin dynamics at MTJs during larval development. Together, these data suggest that specific β-integrin-mediated signals regulate adhesion turnover in response to tension during tissue formation. We propose that integrin–ECM adhesive stability is continuously controlled by force in vivo through integrin-dependent auto-regulatory feedback mechanisms so that tissues can quickly adapt to and withstand mechanical stresses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yuan, L., Fairchild, M. J., Perkins, A. D. & Tanentzapf, G. Analysis of integrin turnover in fly myotendinous junctions. J. Cell Sci. 123, 939–946 (2010).
Gardel, M. L., Schneider, I. C., Aratyn-Schaus, Y. & Waterman, C. M. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26, 315–333 (2010).
Papusheva, E. & Heisenberg, C. P. Spatial organization of adhesion: force-dependent regulation and function in tissue morphogenesis. EMBO J. 29, 2753–2768 (2010).
Parsons, J. T., Horwitz, A. R. & Schwartz, M. A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 (2010).
Ballestrem, C., Hinz, B., Imhof, B. A. & Wehrle-Haller, B. Marching at the front and dragging behind: differential αV β3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 155, 1319–1332 (2001).
Wolfenson, H., Bershadsky, A., Henis, Y. I. & Geiger, B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J. Cell Sci. 124, 1425–1432 (2011).
Schweitzer, R., Zelzer, E. & Volk, T. Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137, 2807–2817 (2010).
Yee, G. H. & Hynes, R. O. A novel, tissue-specific integrin subunit, βν, expressed in the midgut of Drosophila melanogaster. Development 118, 845–858 (1993).
Leptin, M., Bogaert, T., Lehmann, R. & Wilcox, M. The function of PS integrins during Drosophila embryogenesis. Cell 56, 401–408 (1989).
Pines, M., Fairchild, M. J. & Tanentzapf, G. Distinct regulatory mechanisms control integrin adhesive processes during tissue morphogenesis. Dev. Dyn. 240, 36–51 (2011).
Hudson, A. M., Petrella, L. N., Tanaka, A. J. & Cooley, L. Mononuclear muscle cells in Drosophila ovaries revealed by GFP protein traps. Dev. Biol. 314, 329–340 (2008).
Torgler, C. N. et al. Tensin stabilizes integrin adhesive contacts in Drosophila. Dev. Cell 6, 357–369 (2004).
Montana, E. S. & Littleton, J. T. Characterization of a hypercontraction-induced myopathy in Drosophila caused by mutations in Mhc. J. Cell Biol. 164, 1045–1054 (2004).
Sanyal, S., Basole, A. & Krishnan, K. S. Phenotypic interaction between temperature-sensitive paralytic mutants comatose and paralytic suggests a role for N-ethylmaleimide-sensitive fusion factor in synaptic vesicle cycling in Drosophila. J. Neurosci. 19, RC47, 1–5 (1999).
Ezratty, E. J., Bertaux, C., Marcantonio, E. E. & Gundersen, G. G. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J. Cell Biol. 187, 733–747 (2009).
Hynes, R. O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002).
Askari, J. A., Buckley, P. A., Mould, A. P. & Humphries, M. J. Linking integrin conformation to function. J. Cell Sci. 122, 165–170 (2009).
Legate, K. R. & Fassler, R. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 122, 187–198 (2009).
Bajt, M. L. & Loftus, J. C. Mutation of a ligand binding domain of β3 integrin. Integral role of oxygenated residues in α IIb β3 (GPIIb-IIIa) receptor function. J. Biol. Chem. 269, 20913–20919 (1994).
Chen, J., Maeda, T., Sekiguchi, K. & Sheppard, D. Distinct structural requirements for interaction of the integrins α5β1, αvβ5, and αvβ6 with the central cell binding domain in fibronectin. Cell Adhesion Commun. 4, 237–250 (1996).
Jannuzi, A. L., Bunch, T. A., West, R. F. & Brower, D. L. Identification of integrin β subunit mutations that alter heterodimer function in situ. Mol. Biol. Cell 15, 3829–3840 (2004).
Anthis, N. J. et al. Integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J. Biol. Chem. 1–23 (2009).
Datta, A., Huber, F. & Boettiger, D. Phosphorylation of β3 integrin controls ligand binding strength. J. Biol. Chem. 277, 3943–3949 (2002).
Calderwood, D. A. et al. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 277, 21749–21758 (2002).
Tadokoro, S. et al. Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 (2003).
Wegener, K. L. et al. Structural basis of integrin activation by talin. Cell 128, 171–182 (2007).
Tanentzapf, G. & Brown, N. H. An interaction between integrin and the talin FERM domain mediates integrin activation but not linkage to the cytoskeleton. Nat. Cell Biol. 8, 601–606 (2006).
Tanentzapf, G., Martin-Bermudo, M. D., Hicks, M. S. & Brown, N. H. Multiplefactors contribute to integrin-talin interactions in vivo. J. Cell Sci. 119, 1632–1644 (2006).
Ellis, S. J., Pines, M., Fairchild, M. J. & Tanentzapf, G. In vivo functional analysis reveals specific roles for the integrin-binding sites of talin. J. Cell Sci. 124, 1844–1856 (2011).
Harburger, D. S., Bouaouina, M. & Calderwood, D. A. Kindlin-1 and -2 directly bind the C-terminal region of β integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284, 11485–11497 (2009).
Moser, M., Nieswandt, B., Ussar, S., Pozgajova, M. & Fässler, R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14, 325–330 (2008).
Demontis, F. & Perrimon, N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993 (2009).
Press, W. H., Teukolsky, S. A., Vetterling, W. T. & Flannery, B. P. Numerical Recipes: The Art of Scientific Computing 3rd edn (Cambridge Univ. Press, 2007).
Wasserman, L. All of Statistics: A Concise Course in Statistical Inference Ch. 9 (Springer, 2004).
Efron, B. & Tibshirani, R. J. An Introduction to the Bootstrap (Chapman and Hall/CRC, 1994).
Stewart, B. A., Atwood, H. L., Renger, J. J., Wang, J. & Wu, C. F. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A 175, 179–191 (1994).
Franco-Cea, A. et al. Distinct developmental roles for direct and indirect talin-mediated linkage to actin. Dev. Biol. 345, 64–77 (2010).
Acknowledgements
We thank T. Littleton (MIT, USA) and F. Schöck (McGill University, Canada) for flies, and D. Moerman and members of the laboratory for helpful discussions and comments on the manuscript. S.J.E. holds an NSERC Alexander Graham Bell Canada Graduate Scholarship. D.C. is financially supported through an NSERC discovery grant. G.T. is financially supported through NSERC discovery grant 356502 and Canadian Institutes of Health Research operating grant 89835. G.T. is a CIHR New Investigator and Michael Smith Foundation for Health Research Scholar.
Author information
Authors and Affiliations
Contributions
G.T. conceived, designed and supervised the project and participated in data analysis. M.P. carried out most of the genetics crosses, FRAP experiments and data analysis. S.J.E. performed genetic crosses, FRAP experiments and analysed data. L.Y. optimized the conditions and protocols for using the Brkd and para mutants. M.K. performed the force transducer experiments. A.M. and S.C. performed FRAP experiments and analysed data. A.M. carried out the stainings on larval flat preparations. M.P., S.J.E., A.M. and S.C. recorded movies for analysis. R.D and D.C. performed the mathematical modeling and data analysis. G.T., M.P., R.D. and S.J.E. wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1477 kb)
Supplementary Table 1
Supplementary Information (XLSX 18 kb)
Supplementary Movie 1
Supplementary Information (MOV 15173 kb)
Supplementary Movie 2
Supplementary Information (AVI 8632 kb)
Supplementary Movie 3
Supplementary Information (AVI 9800 kb)
Supplementary Movie 4
Supplementary Information (AVI 8209 kb)
Supplementary Movie 5
Supplementary Information (AVI 15430 kb)
Supplementary Movie 6
Supplementary Information (AVI 12255 kb)
Supplementary Movie 7
Supplementary Information (AVI 11134 kb)
Rights and permissions
About this article
Cite this article
Pines, M., Das, R., Ellis, S. et al. Mechanical force regulates integrin turnover in Drosophila in vivo. Nat Cell Biol 14, 935–943 (2012). https://doi.org/10.1038/ncb2555
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2555
This article is cited by
-
Extracellular matrix: an important regulator of cell functions and skeletal muscle development
Cell & Bioscience (2021)
-
Integrin trafficking in cells and tissues
Nature Cell Biology (2019)
-
Multiscale force sensing in development
Nature Cell Biology (2017)
-
A cellular sense of touch
Nature Cell Biology (2012)
-
Unanchoring integrins in focal adhesions
Nature Cell Biology (2012)