Abstract
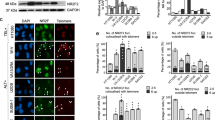
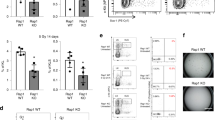
We describe a genome-wide gain-of-function screen for regulators of NF-κB, and identify Rap1 (Trf2IP), as an essential modulator of NF-κB-mediated pathways. NF-κB is induced by ectopic expression of Rap1, whereas its activity is inhibited by Rap1 depletion. In addition to localizing on telomeres, mammalian Rap1 forms a complex with IKKs (IκB kinases), and is crucial for the ability of IKKs to be recruited to, and phosphorylate, the p65 subunit of NF-κB to make it transcriptionally competent. Rap1-mutant mice display defective NF-κB activation and are resistant to endotoxic shock. Furthermore, levels of Rap1 are positively regulated by NF-κB, and human breast cancers with NF-κB hyperactivity show elevated levels of cytoplasmic Rap1. Similar to inhibiting NF-κB, knockdown of Rap1 sensitizes breast cancer cells to apoptosis. These results identify the first cytoplasmic role of Rap1 and provide a mechanism through which it regulates an important signalling cascade in mammals, independent of its ability to regulate telomere function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Ghosh, S. & Karin, M. Missing pieces in the NF-κB puzzle. Cell 109, S81–S96 (2002).
Karin, M. & Ben-Neriah, Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18, 621–663 (2000).
Silverman, N. & Maniatis, T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342 (2001).
Sasaki, Y., Schmidt-Supprian, M., Derudder, E. & Rajewsky, K. Role of NFκB signaling in normal and malignant B cell development. Adv. Exp. Med. Biol. 596, 149–154 (2007).
Pasparakis, M. Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nat. Rev. Immunol. 9, 778–788 (2009).
Aggarwal, B. B. Nuclear factor-κB: the enemy within. Cancer Cell 6, 203–208 (2004).
Tergaonkar, V., Correa, R. G., Ikawa, M. & Verma, I. M. Distinct roles of IκB proteins in regulating constitutive NF-κB activity. Nat. Cell Biol. 7, 921–923 (2005).
Bates, P. W. & Miyamoto, S. Expanded nuclear roles for IκBs. Sci. STKE 2004, pe48 (2004).
Bhoj, V. G. & Chen, Z. J. Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 (2009).
Wertz, I. E & Dixit, V. M. Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 19, 313–324 (2008).
Correa, R. G. et al. Zebrafish IκB kinase 1 negatively regulates NF-κB activity. Curr. Biol. 15, 1291–1295 (2005).
Irelan, J. T. et al. A role for IκB kinase 2 in bipolar spindle assembly. Proc. Natl Acad. Sci. USA 104, 16940–16945 (2007).
Xia, Y. et al. Phosphorylation of p53 by IκB kinase 2 promotes its degradation by β-TrCP. Proc. Natl Acad. Sci. USA (2009).
Chariot, A. The NF-κB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 19, 404–413 (2009).
Perkins, N. D. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25, 6717–6730 (2006).
Moscat, J., Diaz-Meco, M. T. & Wooten, M. W. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32, 95–100 (2007).
Miller, K. M., Ferreira, M. G. & Cooper, J. P. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 24, 3128–3135 (2005).
Brevet, V. et al. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J. 22, 1697–1706 (2003).
Krauskopf, A. & Blackburn, E. H. Rap1 protein regulates telomere turnover in yeast. Proc. Natl Acad. Sci. USA 95, 12486–12491 (1998).
Krauskopf, A. & Blackburn, E. H. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383, 354–357 (1996).
Morse, R. H. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16, 51–53 (2000).
Yang, X., Figueiredo, L. M., Espinal, A., Okubo, E. & Li, B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 137, 99–109 (2009).
Li, B., Oestreich, S. & de Lange, T. Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483 (2000).
de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19, 2100–2110 (2005).
Bianchi, A. & Shore, D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol. Cell 31, 153–165 (2008).
Li, B. & de Lange, T. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 14, 5060–5068 (2003).
Bae, N. S. & Baumann, P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell 26, 323–334 (2007).
Price, C. M. wRAPing up the end to prevent telomere fusions. Mol. Cell 26, 463–464 (2007).
Sfeir, A., Kabir, S., van Overbeek, M., Celli, G. B. & de Lange, T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661 (2010).
O'Sullivan, R. J. & Karlseder, J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11, 171–181 (2010).
Gerhard, D. S. et al. The status, quality, and expansion of the NIH full-length cDNA project: the mammalian gene collection (MGC). Genome Res. 14, 2121–2127 (2004).
Chanda, S. K. et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc. Natl Acad. Sci. USA 100, 12153–12158 (2003).
Celli, G. B. & de Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 (2005).
Chen, L. F. et al. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell Biol. 25, 7966–7975 (2005).
Chen, L. F. & Greene, W. C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 5, 392–401 (2004).
Natoli, G., Saccani, S., Bosisio, D. & Marazzi, I. Interactions of NF-κB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 6, 439–445 (2005).
Kendellen, M. F., Barrientos, K. S. & Counter, C. M. POT1 association with TRF2 regulates telomere length. Mol. Cell Biol. 29, 5611–5619 (2009).
Karlseder, J. et al. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell Biol. 23, 6533–6541 (2003).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Biswas, S. K. et al. Role for MyD88-independent, TRIF pathway in lipid A/TLR4-induced endotoxin tolerance. J. Immunol. 179, 4083–4092 (2007).
Weaver, V. M. et al. β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216 (2002).
Sovak, M. A. et al. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 100, 2952–2960 (1997).
Weigelt, B., Peterse, J. L. & van' t Veer, L. J. Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5, 591–602 (2005).
Basseres, D. S. & Baldwin, A. S. Nuclear factor-κB and inhibitor of κB kinase pathways in oncogenic initiation and progression. Oncogene 25, 6817–6830 (2006).
Dey, A., Wong, E. T., Cheok, C. F., Tergaonkar, V. & Lane, D. P. R-Roscovitine simultaneously targets both the p53 and NF-κB pathways and causes potentiation of apoptosis: implications in cancer therapy. Cell Death Differ. 15, 263–273 (2008).
Dey, A., Wong, E. T., Bist, P., Tergaonkar, V. & Lane, D. P. Nutlin-3 inhibits the NFκB pathway in a p53-dependent manner: implications in lung cancer therapy. Cell Cycle 6, 2178–2185 (2007).
Papa, S. et al. The NF-κB-mediated control of the JNK cascade in the antagonism of programmed cell death in health and disease. Cell Death Differ. 13, 712–729 (2006).
Chua, H. L. et al. NF-κB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 26, 711–724 (2007).
Campbell, M. J. et al. Breast cancer growth prevention by statins. Cancer Res. 66, 8707–8714 (2006).
Biswas, D. K. et al. NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl Acad. Sci. USA 101, 10137–10142 (2004).
Micheau, O. & Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003).
Helbig, G. et al. NF-κB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 278, 21631–21638 (2003).
Rao Ch, V., Li, X., Manna, S. K., Lei, Z. M. & Aggarwal, B. B. Human chorionic gonadotropin decreases proliferation and invasion of breast cancer MCF-7 cells by inhibiting NF-κB and AP-1 activation. J. Biol. Chem. 279, 25503–25510 (2004).
Hagemann, T. et al. Macrophages induce invasiveness of epithelial cancer cells via NF-κB and JNK. J. Immunol. 175, 1197–1205 (2005).
Chew, J. et al. WIP1 phosphatase is a negative regulator of NF-κB signalling. Nat. Cell Biol. 11, 659–666 (2009).
Park, J. I. et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72 (2009).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Tergaonkar, V., Bottero, V., Ikawa, M., Li, Q. & Verma, I. M. IκB kinase-independent IκBα degradation pathway: functional NF-κB activity and implications for cancer therapy. Mol. Cell Biol. 23, 8070–8083 (2003).
Deng, Z., Atanasiu, C., Burg, J. S., Broccoli, D. & Lieberman, P. M. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 77, 11992–12001 (2003).
Cho, C. Y. et al. Identification of the tyrosine phosphatase PTP-MEG2 as an antagonist of hepatic insulin signaling. Cell Metab. 3, 367–378 (2006).
Luesch, H. et al. A functional genomics approach to the mode of action of apratoxin A. Nat. Chem. Biol. 2, 158–167 (2006).
Yu, F. et al. Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog. 3, e44 (2007).
Acknowledgements
We wish to thank the Agency for Science Technology and Research, Singapore (A*Star) for funding and support to the V.T. laboratory. I.M.V. is an American Cancer Society Professor of Molecular Biology, and holds the Joan and Irwin Jacobs Chair in Exemplary Life Sciences. This work was supported in part by grants from the Leducq Foundation, Meriaux Foundation, Ellison Medical Foundation, Ipsen/Biomeasure, Sanofi Aventis, and the H.N. and Frances C. Berger Foundation. We thank T. Murphy and N. Tonnu for help with the intial investigations involved in this work. We are grateful to B. Li and T. De Lange for Rap1 deletion constructs and P. Lieberman for the Rap1 shRNA vector. We thank J. Karlseder for helpful discussions.
Author information
Authors and Affiliations
Contributions
H.T., A.G., E.T.W., N.M., M.W. and V.T. carried out biochemical experiments with Rap1, generated Rap1-mutant mice and helped in data analysis. S.G., A.S.P., J.D.O., N.L.T. and L.J.S. carried out the analysis of Rap1 in human cancers and also helped in data analysis. H.L., P.D.J., A.O., E.S., P.S., S.K.C. and V.T. carried out the genome-wide screen and helped in data analysis and initial characterization of Rap1 as an NF-κB regulator. S.K.C., I.M.V. and V.T. did the project planning and data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1112 kb)
Rights and permissions
About this article
Cite this article
Teo, H., Ghosh, S., Luesch, H. et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-κB-dependent gene expression. Nat Cell Biol 12, 758–767 (2010). https://doi.org/10.1038/ncb2080
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2080
This article is cited by
-
ZNF524 directly interacts with telomeric DNA and supports telomere integrity
Nature Communications (2023)
-
Anesthetic propofol enhances cisplatin-sensitivity of non-small cell lung cancer cells through N6-methyladenosine-dependently regulating the miR-486-5p/RAP1-NF-κB axis
BMC Cancer (2022)
-
The telomere complex and the origin of the cancer stem cell
Biomarker Research (2021)
-
Non-canonical roles of canonical telomere binding proteins in cancers
Cellular and Molecular Life Sciences (2021)
-
The role of telomere-binding modulators in pluripotent stem cells
Protein & Cell (2020)