Abstract
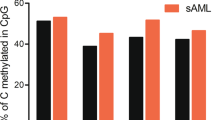
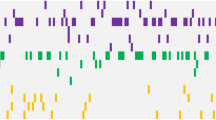
Alterations in DNA methylation have been implicated in the pathogenesis of myelodysplastic syndromes (MDS), although the underlying mechanism remains largely unknown. Methylation of CpG dinucleotides is mediated by DNA methyltransferases, including DNMT1, DNMT3A and DNMT3B. DNMT3A mutations have recently been reported in patients with de novo acute myeloid leukemia (AML), providing a rationale for examining the status of DNMT3A in MDS samples. In this study, we report the frequency of DNMT3A mutations in patients with de novo MDS, and their association with secondary AML. We sequenced all coding exons of DNMT3A using DNA from bone marrow and paired normal cells from 150 patients with MDS and identified 13 heterozygous mutations with predicted translational consequences in 12/150 patients (8.0%). Amino acid R882, located in the methyltransferase domain of DNMT3A, was the most common mutation site, accounting for 4/13 mutations. DNMT3A mutations were expressed in the majority of cells in all tested mutant samples regardless of myeloblast counts, suggesting that DNMT3A mutations occur early in the course of MDS. Patients with DNMT3A mutations had worse overall survival compared with patients without DNMT3A mutations (P=0.005) and more rapid progression to AML (P=0.007), suggesting that DNMT3A mutation status may have prognostic value in de novo MDS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Esteller M . Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159.
Sharma S, Kelly TK, Jones PA . Epigenetics in cancer. Carcinogenesis 2009; 31: 27–36.
Ting AH, McGarvey KM, Baylin SB . The cancer epigenome--components and functional correlates. Genes Dev 2006; 20: 3215–3231.
Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010; 17: 13–27.
Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood 2009; 114: 3448–3458.
Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O’Keefe C et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood 2009; 113: 1315–1325.
Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF . Multicenter phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol 2009; 28: 556–561.
Chitambar CR, Libnoch JA, Matthaeus WG, Ash RC, Ritch PS, Anderson T . Evaluation of continuous infusion low-dose 5-azacytidine in the treatment of myelodysplastic syndromes. Am J Hematol 1991; 37: 100–104.
Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006; 106: 1794–1803.
Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol 2002; 20: 2429–2440.
Goll MG, Bestor TH . Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74: 481–514.
Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW et al. Induction of tumors in mice by genomic hypomethylation. Science 2003; 300: 489–492.
Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 1995; 81: 197–205.
Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999; 402: 187–191.
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363: 2424–2433.
Graubert TA, Payton MA, Shao J, Walgren RA, Monahan RS, Frater JL et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS One 2009; 4: e4583.
Ng PC, Henikoff S . Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874.
Ramensky V, Bork P, Sunyaev S . Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900.
Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL et al. Array-based genomic resequencing of human leukemia. Oncogene 2010; 29: 3723–3731.
Gowher H, Loutchanwoot P, Vorobjeva O, Handa V, Jurkowska RZ, Jurkowski TP et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol 2006; 357: 928–941.
Okano M, Bell DW, Haber DA, Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257.
Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004; 429: 900–903.
Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H . De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med 2007; 204: 715–722.
Subramanyam D, Belair CD, Barry-Holson KQ, Lin H, Kogan SC, Passegue E et al. PML-RAR{alpha} and Dnmt3a1 cooperate in vivo to promote acute promyelocytic leukemia. Cancer Res 2010; 70: 8792–8801.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 2010; 42: 722–726.
Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 2009; 113: 6403–6410.
Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 2009; 41: 838–842.
Mohamedali AM, Smith AE, Gaken J, Lea NC, Mian SA, Westwood NB et al. Novel TET2 mutations associated with UPD4q24 in myelodysplastic syndrome. J Clin Oncol 2009; 27: 4002–4006.
Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 2010; 42: 665–667.
Tefferi A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Patnaik MM et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia 2009; 23: 1343–1345.
Viguie F, Aboura A, Bouscary D, Ramond S, Delmer A, Tachdjian G et al. Common 4q24 deletion in four cases of hematopoietic malignancy: early stem cell involvement? Leukemia 2005; 19: 1411–1415.
Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science (New York, NY 2007; 318: 447–450.
Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at Scerevisiae telomeres. Embo J 1997; 16: 3219–3232.
Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010; 468: 839–843.
Acquaviva C, Gelsi-Boyer V, Birnbaum D . Myelodysplastic syndromes: lost between two states? Leukemia 2010; 24: 1–5.
Sinclair DA, Milne TA, Hodgson JW, Shellard J, Salinas CA, Kyba M et al. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development 1998; 125: 1207–1216.
Steensma DP, Higgs DR, Fisher CA, Gibbons RJ . Acquired somatic ATRX mutations in myelodysplastic syndrome associated with alpha thalassemia (ATMDS) convey a more severe hematologic phenotype than germline ATRX mutations. Blood 2004; 103: 2019–2026.
Acknowledgements
This work was supported by NIH grants R01HL082973 (Graubert), RC2HL102927 (Graubert), U54HG003079 (Wilson), P01CA101937 (Ley), and a Howard Hughes Medical Institute Physician-Scientist Early Career Award (Walter).
Author Contributions
MW, LD, TL and TG designed the study. DS, JS, MG, MM, RF, HS, JKV and MO performed research and generated data. MW, LD, DS, JS, JB, PW, JD, EM, RW, TL and TG analyzed data. MW, LD, DS and TG wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
These authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Walter, M., Ding, L., Shen, D. et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25, 1153–1158 (2011). https://doi.org/10.1038/leu.2011.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2011.44
Keywords
This article is cited by
-
Clonal haematopoiesis - a novel entity that modifies pathological processes in elderly
Cell Death Discovery (2023)
-
Genomic characterization of IDH-mutant astrocytoma progression to grade 4 in the treatment setting
Acta Neuropathologica Communications (2023)
-
The methyltransferase domain of DNMT1 is an essential domain in acute myeloid leukemia independent of DNMT3A mutation
Communications Biology (2022)
-
Predictive values of mutational variant allele frequency in overall survival and leukemic progression of myelodysplastic syndromes
Journal of Cancer Research and Clinical Oncology (2022)
-
DNA methylation: a saga of genome maintenance in hematological perspective
Human Cell (2022)