Abstract
The 621+3 A>G variant of the CFTR gene was initially detected in four Greek patients with a severe form of cystic fibrosis, and it is reported to impair CFTR mRNA splicing. We present three lines of evidence that argue against the pathogenicity of this variant. First, its allelic frequency in the Italian population was 0.4%. Even considering the lowest value in the confidence interval we would expect 10% of Italian CF patients to be heterozygotes for this variant, whereas it has been reported only in one patient (0.04% of Italian CF patients). Second, expression of the 621+3 A>G variant in HeLa cells using a hybrid minigene showed that 39.5±1.1% of transcripts were correctly spliced, indicating that its effects on mRNA splicing are similar to those of the CFTR intron 8 5T variant, associated with congenital bilateral absence of vas deferens (CBAVD), but not with CF. Third, we have identified an asymptomatic individual who harbored the 621+3 A>G variant in trans with the Q552X mutation. Because 621+3 A>G is often included in population-screening programs, this information is critical to provide adequate counseling to patients. Further work should be aimed at investigating whether this variant may have a role in CBAVD or atypical CF.
Similar content being viewed by others
Introduction
Cystic fibrosis (CF; MIM no. 219 700) is one of the most frequent autosomal recessive disorder in the Caucasian population, affecting 1:2500 individuals and it is caused by mutations of the CFTR gene.1 The frequency of heterozygous carriers in the European population is about 1 in 25 individuals; therefore several countries have instituted specific population-screening programs to identify carriers. Because more than 1600 different CFTR mutations have been reported in literature, these programs usually provide testing for a panel which includes the 20–50 most common CFTR mutations in that particular population.2 However, the pathogenicity of some mutations included in these panels has been disputed. An example is the I148T variant initially reported as a CF mutation, which is now considered a non-pathogenic nucleotide change.3
We have chosen to study the 621+3 A>G nucleotide change because one of us had tested positive for this variant during a CF-screening program. The 621+3 A>G variant was first reported to the Cystic Fibrosis Mutation Database (http://www.genet.sickkids.on.ca/cftr/app) as a neutral polymorphism. Subsequently, it was described as a severe disease-causing mutation in four CF patients. It was reported to affect the splicing of exon 4, leading to the formation of a non-functional CFTR protein.4 We now present epidemiological, functional and clinical evidence suggesting that the 621+3 A>G variant should not be considered a CF disease-causing mutation.
Materials and methods
Analysis of 621+3 A>G variant in the general population
We studied a cohort of 250 healthy individuals of northern Italian origin, aged 20–45 years. The screening of the 621+3 A>G variant has been performed by High Resolution Melting (HRM) analysis using the Rotorgene 6000 (Corbett Life Sciences, Concorde, NSW, Australia). PCR primers were: FOR 5′-CACATTGGAATGCAGATGAGA-3′ and REV 5′-TCCCTTACTTGTACCAGCTCACT-3′; amplification conditions were 95 °C 5 min, 50 cycles of 95 °C 10 s, 60 °C 20 s and a final step of 72 °C 5 min. HRM analysis was performed between 71 and 83 °C. One 621 +3 A>G heterozygote and one wild-type homozygote were used as controls. All samples showing the HRM profile of the heterozygote have been sequenced using the amplification primers.
Expected frequencies of CF genotypes in the population were calculated using the Hardy–Weinberg equation.5 95% confidence interval (CI) of the proportion was calculated as described.6
Generation of the minigene construct
To obtain the backbone of the artificial minigene, a 1.8 kb fragment encompassing the entire human beta-globin gene was amplified from genomic DNA of a healthy volunteer using primers FOR 5′-CTTCTGCTAGCAGTCAGGGCAGAGCCATCTA-3′ REV 5′-CTTCTGGGCCCTTTGCAGCCTCACCTTCTTT-3′ (which introduce NheI and ApaI sites in the product) and the Expand Long Template PCR System (Roche Diagnostics GmbH, Mannhein, Germany). PCR conditions were 94 °C 2 min 15 cycles of 94 °C 10 s, 65° C 30 s, 68 °C 3 min, followed by 20 cycles of 94 °C 15 s, 65° C 30 s, 68 °C 3 min, with a 15 s increase every cycle, and a final extension step of 68 °C 7 min.
After enzymatic digestion it was cloned into the corresponding sites of pCDNA3.1 (Invitrogen, Carlsbad, CA, USA).
An artificial multiple cloning site containing the XhoI, NotI and HindIII sites (in capital letters in the following sequence) tgtacaCTCGAGttatGCGGCCGCtactAAGCTTGTACA was then cloned into the BsrGI site within intron 2 of the β-globin gene.
A 853 bp fragment encompassing 341 bp of intron 3, exon 4 and 296 bp of intron 4 of the CFTR gene was amplified from genomic DNA of a healthy individual identified as carrier of the 621+3 A>G through population screening, using primers: FOR 5′-CTTCTCTCGAGAAAAGGGAAATGCTTTAGAAACTG-3′ and REV 5′-CTTCTAAGCTTTGAGTCATCTTAACAGGAAACCA-3′ (which introduce XhoI and HindIII restriction sites in the PCR product) and Applied Biosystems AmpliTaq Gold DNA Polymerase. PCR conditions were: 94 °C 10 min, 30 cycles of 95 °C 30 s, 60° C 40 s, 72 °C 1 min and a final extension step 72 °C 10 min. After digestion the fragment was cloned into the minigene vector, and sequenced. One clone containing the wild-type allele, and one with the mutation were retained for expression experiments.
Minigene expression and analysis of transcripts
HeLa cells (2x105) were transfected with 400 ng of the wild type or the mutated minigene using Effectene reagent (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instruction. After Total RNA was extracted and retrotranscribed as reported7 using primers FOR 5′-TCTGTCCACTCCTGATGCTG-3′ and REV 5′-CACTGGTGGGGTGAATTCTT-3′ located on β-globin exon 2 and 3 and Roche Taq DNA polymerase. PCR conditions were: 94 °C 3 min, 30 cycles of 94 °C 1 min, 55 °C 1 min, 72 °C 45 s and a final extension step 72 °C 7 min. This set of primers does not amplify eventual ectopically expressed CFTR transcripts.
PCR products were separated either on a 3% agarose gel or on a 12% acrilamyde gel. Individual bands were excised and sequenced using amplification primers as described.8 PCR products were also labeled with α32P-dATP using the “last cycle hot” procedure9 and after gel separation were visualized and quantitated using a Storm Phosporimager (Molecular Dynamics, Sunnyvale, CA, USA). The quantitation was performed on three independent experiments.
Results
Epidemiology
Two out of 250 normal individuals (500 alleles) were heterozygous for the 621+3 A>G mutation giving an allele frequency of 0.4%, 95% CI (0.11–1.45). Figure 1 shows the abnormal HRM profile in these individuals.
Based on these data the expected frequency of individuals harboring the 621+3 A>G allele in compound heterozygosity with another CF allele would be 1:7812 (30% of CF patients), whereas the frequency of homozygotes would be 1:62500 (4% of CF patients). Even considering the lowest value in the 95% CI the expected frequencies would be 1:24010 (10% of CF patients) for heterozygotes, and 1:800000 (0.3% of CF patients) for homozygotes.
The Italian CFTR mutation database reports only one patient who was compound heterozygous for the 621+3A>G allele out of a total of 2253 affected individuals (4506 alleles) (http://spazioinwind.libero.it/laboratoriCF/frequenze.htm).
Molecular studies
Because epidemiological data were arguing against the pathogenicity of the 621+3 A>G, we investigated CFTR mRNA splicing using a minigene-based approach.
Constructs containing wild-type and mutant CFTR exon 4 and the intron–exon boundaries were cloned into the pcDNA3.1-β-globin construct and expressed in HeLa cells. mRNA was analyzed by RT-PCR using β-globin-specific primers (Figure 2a), that do not amplify eventual ectopically expressed CFTR transcripts. We also did not find evidence of expression of the endogenous β-globin gene (not shown).
(a) Structure of the hybrid minigene (see Materials and methods section for details). The arrows indicate the primers used for RT-PCR analysis. Boxed in gray, β-globin exons 1–3, in white CFTR exon 4. Expression is driven by the CMV promoter of pcDNA3.1. (b) RT-PCR analysis of mRNA of cell expressing the wild-type (WT) minigene, or the construct harboring the 621+3 A>G variant (MUT). Fragments were labeled with 32P using the last cycle hot procedure (see Materials and methods section for details) and visualized with a phosphor-Imager. On the right the composition of each transcript. The experiment was performed in triplicate. (c) Densitometric profile above the gel. Arrows indicate the peaks corresponding to each fragment. Quantitation of three independent experiments revealed that fragment 1 represents 39.4±1.1% of total transcripts, fragment 2 58.5±0.8% and fragment 3 2.0±0.4%.
As seen in Figure 2b the transcript derived from the wild-type construct is correctly spliced, producing a single transcript containing the entire CFTR exon 4. Instead, the mutated construct yielded three different bands, one corresponding to the correctly spliced transcript, one resulting from the activation of a cryptic splice site within exon 4, 93 bp upstream of the exon 4/intron 4 boundary, and a faint third band corresponding to a transcript lacking the entire CFTR exon 4. Although these transcripts maintain the reading frame of the mRNA, the resulting proteins are likely to be non-functional.10 Relative quantification of radiolabelled RT-PC products (Figure 2c) was performed on three independent experiments and revealed that the aberrantly spliced forms account for 58.5±0.8% (transcript 2 in Figure 1b) and 2.0±0.4% (transcript 3 in Figure 2b), whereas the correctly spliced product accounts for 39.4±1.1% of total transcripts (transcript 1 in Figure 2b).
Patient data
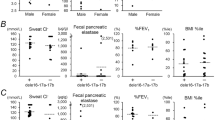
In the course of the study we identified by chance an individual who harbored the 621 +3A>G variant in trans with the pathogenic Q552X-truncating mutation. She is an 8-year-old girl, first child of healthy unrelated parents. Immunoreactive trypsinogen at birth was normal. She had an unremarkable medical history in particular; she never experienced severe respiratory infections, episodes of wheezing, sinusitis or recurrent diarrhea. Stature and weight were both above the 75th centile. She was identified to be the carrier of both alterations because her parents underwent CF screening during a second pregnancy: the mother was found to harbor 621+3 A>G, whereas the father had tested positive for Q552X. After the molecular diagnosis she underwent a thorough medical evaluation. Chest X-Ray was normal, serum amylase, lipase and fecal chymotrypsine levels were all within normal ranges. Chloride sweat test was repeated three times. In one occasion chloride sweat test was pathologic (75 mEq/l), and in two occasions it yielded borderline results (49 mEq/l).
Discussion
In Southern European populations the proportion of non F508del alleles in CF carriers is high,11 but the pathogenicity of some of these variants is disputed. Defining the exact role of these nucleotide substitutions is essential to provide genetic counseling to subjects identified through population screening.
The 621+3 A>G variant has been reported in Greek patients with severe form of CF. We now present three lines of evidence that argue against the pathogenicity of the 621+3 A>G mutation. The first set of data comes from epidemiological analysis. We observed that this variant is more frequent in the general population than in patients. Its allelic frequency in our sample of healthy Italian controls was 0.4% (CI 95% 0.11–1.45), making it one of the most frequent CF alleles. In other words, even considering the lowest value in the 95% CI, we would have expected to find at least 225 patients with this mutation in the Italian CF registry. In contrast, it was reported only in 1/2253 CF patients. It should be noted that in most Italian centers mutation screening in CF patients is performed by denaturing gradient gel electrophoresis or DHPLC, therefore this mutation should have been identified if present.
The second line of evidence derives from our minigene experiments. Our data clearly show that although this nucleotide change does affect mRNA processing, a considerable portion of transcripts (39.4±1.1%) are correctly spliced, thus giving rise to functional CFTR proteins. Another mutation targets the same splice site in position +1 and does not allow formation of any residual correctly spliced transcripts.10 Interestingly, the 621+1 G>T mutation causes the activation of the same cryptic splice within exon 4. It should be noted that position +3 in the 5′ splice site is less critical than positions +1 or +2, with both A and G nucleotides being almost equally represented in this position,12 therefore it is not surprising that 621+3 A>G has a less detrimental effect on mRNA splicing than 621+1 G>T. Although mRNA splicing efficiency can vary between different tissues,13 these data suggest that the effect of 621+3 A>G on mRNA splicing is similar to that of the CFTR intron 8 5 T variant,14 involved in congenital bilateral absence of vas deferens, but not in CF.
Our data are not in contrast with the results obtained by Tzetis et al.4 In fact, we identified the same splicing alteration qualitatively. The discrepancy in the conclusions lies in the fact that methodology employed by Tzetis et al.4 could not discriminate the relative contribution of each individual allele to the total pool of correctly spliced transcripts, and therefore they could not rule out the presence of correctly spliced transcripts deriving from the 621+3 A>G allele. In fact, traditional RT-PCR analysis of patient mRNA cannot distinguish transcripts deriving from individual alleles of compound heterozygotes (as were the four reported 621+3 A>G patients), unless the analysis is coupled to either selective digestion of one of the two alleles by a specific restriction endonuclease,15 or if the amplification protocol is allele-specific.14 This type of analysis is not always possible because of the absence of informative polymorphic sites in the transcript. Hybrid minigenes overcome these limitations, and in fact they have been widely used to study CFTR-splicing alterations.16, 17, 18
Finally, we have identified an asymptomatic 8-year-old girl harboring 621+3 A>G allele in trans with a truncating CFTR mutation. Although she had a borderline sweat test she did not present with any symptom of CF. It will be important to monitor the patient to determine if in the future she will develop an atypical form of CF, because the quantity of correctly spliced transcripts may differ among various organs of the same patient, contributing to differential organ disease manifestations.
A likely explanation of the severe phenotype of the four reported patients (and of the single Italian patient with harboring 621+3 A>G) is that another undetected mutation was in cis with 621+3 A>G. A complex allele comprising 621+3 A>G and a 4332delTG has been identified in a patient of north-African origin.19 It is likely that the mutation analysis protocol, carried out by denaturing gradient gel electrophoresis, would have identified this mutation (if present) in the four Greek patients with 621+3 A>G (in any case we ruled out the presence of this variant in all individuals carrying the 621+3 A>G allele included in our study). However, this technique cannot detect heterozygous deletions of individual exons, that may be present in up to 60% of classic CF patients in whom a single-point mutation is detected.20 This case is similar to that of the I148T variant, which by itself is not a CF disease-causing mutation, but in some patients it is found in cis with the 3199del6 mutation.21 Extensive analysis of CFTR, including gene rearrangements, should be performed in all patients with typical CF symptoms harboring the 621+3 A>G allele, to identify a possible other mutation in cis with this variant, which could account for the severe phenotype.
In conclusion, our data suggest that 621+3 A>G should not be considered a severe CF mutation because of its frequency in the population, because it still allows the synthesis of significant amounts of functional CFTR protein, and the absence of clinical disease in a compound (621+3 A>G/ Q552X) heterozygote individual. Although a large study performed in the Italian population did not detect the presence of this mutation in infertile men,22 further epidemiological studies should be aimed at determining whether 621+3 A>G in isolation may have a role in congenital bilateral absence of vas deferens or in atypical forms of CF.
References
Davies, J. C., Alton, E. W. & Bush, A. Cystic fibrosis. BMJ 335, 1255–1259 (2007).
Tomaiuolo, R., Spina, M. & Castaldo, G. Molecular diagnosis of cystic fibrosis: comparison of four analytical procedures. Clin Chem Lab Med 41, 26–32 (2003).
Claustres, M., Altieri, J. P., Guittard, C., Templin, C., Chevalier-Porst, F. & Des Georges, M. Are p.I148T, p.R74W and p.D1270N cystic fibrosis causing mutations? BMC Med Genet 5, 19 (2004).
Tzetis, M., Efthymiadou, A., Doudounakis, S. & Kanavakis, E. Qualitative and quantitative analysis of mRNA associated with four putative splicing mutations (621+3A–>G, 2751+2T–>A, 296+1G–>C, 1717-9T–>C-D565G) and one nonsense mutation (E822X) in the CFTR gene. Hum Genet 109, 592–601 (2001).
Hardy, G. H. Mendelian Proportions in a Mixed Population. Science 28, 49–50 (1908).
Newcombe, R. G. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med 17, 857–872 (1998).
Trevisson, E., Salviati, L., Baldoin, M. C., Toldo, I., Casarin, A., Sacconi, S. et al. Argininosuccinate lyase deficiency: mutational spectrum in Italian patients and identification of a novel ASL pseudogene. Hum Mutat 28, 694–702 (2007).
Salviati, L., Trevisson, E., Baldoin, M. C., Toldo, I., Sartori, S., Calderone, M. et al. A novel deletion in the GJA12 gene causes Pelizaeus-Merzbacher-like disease. Neurogenetics 8, 57–60 (2007).
Sacconi, S., Salviati, L., Nishigaki, Y., Walker, W. F., Hernandez-Rosa, E., Trevisson, E. et al. A functionally dominant mitochondrial DNA mutation. Hum Mol Genet 17, 1814–1820 (2008).
Zielenski, J., Bozon, D., Markiewicz, D., Aubin, G., Simard, F., Rommens, J. M. et al. Analysis of CFTR transcripts in nasal epithelial cells and lymphoblasts of a cystic fibrosis patient with 621+1G–>T and 711+1G–>T mutations. Hum Mol Genet 2, 683–687 (1993).
Chillon, M., Casals, T., Gimenez, J., Ramos, M. D., Palacio, A., Morral, N. et al. Analysis of the CFTR gene confirms the high genetic heterogeneity of the Spanish population: 43 mutations account for only 78% of CF chromosomes. Hum Genet 93, 447–451 (1994).
Cartegni, L., Chew, S. L. & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3, 285–298 (2002).
Mak, V., Jarvi, K. A., Zielenski, J., Durie, P. & Tsui, L. C. Higher proportion of intact exon 9 CFTR mRNA in nasal epithelium compared with vas deferens. Hum Mol Genet 6, 2099–2107 (1997).
Rave-Harel, N., Kerem, E., Nissim-Rafinia, M., Madjar, I., Goshen, R., Augarten, A. et al. The molecular basis of partial penetrance of splicing mutations in cystic fibrosis. Am J Hum Genet 60, 87–94 (1997).
Martella, M., Salviati, L., Casarin, A., Trevisson, E., Opocher, G., Polli, R. et al. Molecular analysis of two uncharacterized sequence variants of the VHL gene. J Hum Genet 51, 964–968 (2006).
Zuccato, E., Buratti, E., Stuani, C., Baralle, F. E. & Pagani, F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J Biol Chem 279, 16980–16988 (2004).
Pagani, F., Stuani, C., Zuccato, E., Kornblihtt, A. R. & Baralle, F. E. Promoter architecture modulates CFTR exon 9 skipping. J Biol Chem 278, 1511–1517 (2003).
Hefferon, T. W., Groman, J. D., Yurk, C. E. & Cutting, G. R. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci USA 101, 3504–3509 (2004).
Loumi, O., Ferec, C., Mercier, B., Creff, J., Fercot, B., Denine, R. et al. CFTR mutations in the Algerian population. J Cyst Fibros 7, 54–59 (2008).
Hantash, F. M., Redman, J. B., Starn, K., Anderson, B., Buller, A. & McGinniss, M. J. et al. Novel and recurrent rearrangements in the CFTR gene: clinical and laboratory implications for cystic fibrosis screening. Hum Genet 119, 126–136 (2006).
Monaghan, K. G., Highsmith, W. E., Amos, J., Pratt, V. M., Roa, B., Friez, M. et al. Genotype-phenotype correlation and frequency of the 3199del6 cystic fibrosis mutation among I148T carriers: results from a collaborative study. Genet Med 6, 421–425 (2004).
Morea, A., Cameran, M., Rebuffi, A. G., Marzenta, D., Marangon, O., Picci, L. et al. Gender-sensitive association of CFTR gene mutations and 5T allele emerging from a large survey on infertility. Mol Hum Reprod 11, 607–614 (2005).
Acknowledgements
Dr Salviati is supported by a grant from AFM and by Telethon Italia Grant GGP06256.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Forzan, M., Salviati, L., Pertegato, V. et al. Is CFTR 621+3 A>G a cystic fibrosis causing mutation?. J Hum Genet 55, 23–26 (2010). https://doi.org/10.1038/jhg.2009.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.115
Keywords
This article is cited by
-
Demographic characteristics, clinical and laboratory features, and the distribution of pathogenic variants in the CFTR gene in the Cypriot cystic fibrosis (CF) population demonstrate the utility of a national CF patient registry
Orphanet Journal of Rare Diseases (2021)
-
Parent-of-origin effect of hypomorphic pathogenic variants and somatic mosaicism impact on phenotypic expression of retinoblastoma
European Journal of Human Genetics (2018)
-
A synonymous splicing mutation in the SF3B4 gene segregates in a family with highly variable Nager syndrome
European Journal of Human Genetics (2017)