Abstract
Frequent occurrence of [4+2] adducts in the secondary metabolites suggested involvement of Diels–Alderases (DAases) in their biosynthesis. However, a limited number of DAases were reported before early 2000s. Advancements in whole-genome sequencing and searching tool of the biosynthetic gene clusters of the secondary metabolites facilitate the identification of plausible DAases. Thus, during past 5 years, nine DAases have been characterized by genetic and biochemical analyses. These include a detailed functional analysis of SpnF that solely catalyzes [4+2] cycloaddition, a structural analysis of spirotetramate-forming enzyme PyrI4 complexed with the corresponding cycloadduct, and DAases catalyzing decalin formation and macrocyclic pyridine formation. Together with decalin-forming enzymes and macrocyclic pyridine-forming enzymes, these results provided sufficient data to discuss catalytic mechanism of DAases and nature’s strategy for molecular diversification of linear chain intermediates derived from polyketide and ribosomal peptide biosynthetic machinery.
Similar content being viewed by others
Introduction
The Diels–Alder reaction is classified as a [4+2] cycloaddition in the pericyclic reaction involving a 1,3-diene and an alkene (dienophile) to afford a six-membered ring with four contiguous stereocenters in highly regio- and stereospecific manners. Due to these exquisite selectivities, the Diels–Alder reaction has been utilized for the synthesis of structurally intricate natural products with pharmaceutical and biological activities.1, 2 In principle, Diels–Alder reaction proceeds in a concerted manner. Along this line, Williams and co-workers3 discussed concertedness of Diels–Alder reactions in both enzymatic and non-enzymatic reactions. However, regardless confirming its mechanism, synthetic chemists frequently call this extremely useful cyclization reaction as ‘a formal Diels–Alder reaction’. Extensive survey of natural formal [4+2] adducts suggested that >400 natural products are potentially biosynthesized by formal Diels–Alderases (DAases).4, 5, 6 Representative examples of natural [4+2] cycloadducts are shown in Figure 1. These include intermolecular adducts (plagiospirolide A, obtunone and guaiane sesquiterpene dimer),7, 8, 9 intramolecular adducts (ircinianin, chaetoglobosin A)10, 11 and a hetero-Diels–Alder adducts (grandione).12
During late 1990s to early 2000s, three DAases, solanapyrone (1) synthase (Sol5: oxidase),13 lovastatin (2) nonaketide synthase (LovB: polyketide synthase (PKS))14 and macrophomate (3) synthase (MPS)15, 16 were reported. The former two enzymes catalyzed intramolecular reactions yielding decalin scaffolds while the latter DAase catalyzed intermolecular reaction. These enzymes catalyzed formation of reactive diene/dienophile substrates and further catalyzed the corresponding [4+2] cycloaddition (Schemes 1 and 2).5, 6 The bifunctional DAases, Sol5, LovB and MPS were not categorized as a single enzyme family because they did not share any common catalytic mechanism. Characterization of these enzymes required significant efforts such as purification of enzymes and synthetic preparation of all putative substrate candidates. However, advancements in whole-genome sequencing and searching tool of biosynthetic gene clusters of the secondary metabolites facilitate the identification of plausible DAases. In fact, for recent 5 years, more than nine DAases catalyzing a formal intramolecular [4+2] cycloadditions have been identified in the biosynthesis of polyketides and ribosomal peptides. In this review, we hope to highlight those enzymes and discuss the mechanism of their catalyses.
Intramolecular DAases generating decalin skeletons
PKSs, particularly bacterial multimodular and fungal iterative PKSs, generate various skipped polyene intermediates with all E-configurations.17 Among them, the polyene chains interrupted by four contiguous sp3 carbons provide suitable triene system for intramolecular [4+2] cycloaddition. Due to frequent occurrence of this triene system, decalin skeleton is one of the most abundant [4+2] adducts among polyketides. Feasiblity of this hypothesis is supported by corresponding biomimetic total synthesis as in the case of UCS1025A.18
To the bifunctional DAases, Sol5 and LovB, catalyzing decalin formation, a new member has joined recently. Betaenones B (4) is phytotoxic polyketide isolated from Phoma betae Fr.19, 20 In 2015, we identified a betaenone octaketide synthase (Bet1) and a trans-acting enoyl reductase (Bet3), which participate in the formation of dehydroprobetaenone I (5) having a decalin scaffold by heterologous expression of the corresponding genes (Scheme 3).21 Chemical reactivity and conformational analysis suggested that involvement of DAase PKS Bet1 at the stage of reductive release of growing polyketide chain giving reactive trienone although non-enzymatic reaction22 cannot be ruled out.
During the biosynthetic study of fungal metabolite fusarisetin A (6),23 gene deletion experiments of fsa2, a functionally unknown protein, gave a diastereomeric mixture of putative Diels–Alder adducts (Scheme 3).24 Considering the wild-type strain yielded only an endo-adduct, it was proposed that the reactive tetraene precursor gave a mixture of isomerric adducts spontaneously in this mutant. Due to absence of other enzymatic activity, Fsa2 was regarded as a monofunctional DAase to catalyze the endo-selective [4+2] cycloaddition. Intriguingly, the essentially same result on a homologous CghA has been reported in the biosynthesis of other tetramate decalin metabolite Sch210972 (7).25, 26 Computational analysis of the [4+2] cycloaddition with a truncated substrate indicated that the enzyme accelerates the cycloaddition a 1000-fold at 30 °C.25, 26 Homologous genes are also found in the biosynthetic gene clusters of the tetramic acid-containing adducts such as equisetin (Eqx3; 90% identity)27 and pyrrolocin (gNR600; 37%)28 and macrocyclic adduct cytochalasin (CcsF; 27%).29 Frequent occurrence of DAase genes with PKS-NRPS genes suggested co-evolution of these genes.
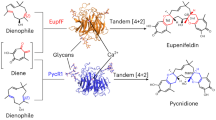
The second group of monofunctional Diels–Alderase genes was identified in the biosynthetic gene cluster of pyrroindomycin A (8),30, 31 a bacterial spirotetronate/spirotatramate family member possessing a tetramate decalin skeleton. Comparative analysis of the gene clusters revealed PyrE3, which belongs to a flavoenzyme superfamily, as a potential candidate for the [4+2] cycloaddition.32 Gene deletion and in vitro experiments showed that PyrE3 catalyzed the endo-selective cycloaddition of a linear polyene precursor 9 after cleaving from the PKS (Scheme 4). Although the recombinant PyrE3 has a flavin adenine diphosphate (FAD) in a non-covalent manner, PyrE3 did not catalyze the redox reaction and solely catalyzed the cycloaddition. Interestingly, a FAD-deficient mutant showed no catalytic activity, suggesting essential role of the FAD to maintain the requisite arrangement of the active site for the endo-selective cycloaddition.32 Currently, five gene clusters of this family member versipelostatin (12),33 chlorothricin (13),34 kijanimicin35 and tetrocarcin36 have pyrE3 homologs (vstK, kijA, chlE3 and tcaE1, 43–52% identity). Another Diels–Alderase, PyrI4, found in the same biosynthetic gene cluster is discussed in the next section.
Previously, enzymatic decalin scaffold formations by [4+2] cycloaddition were firmly established in the biosyntheses of the two fungal polyketides, solanapyrone13, 37 and lovastatin14, 38 (Scheme 1). The former oxidase Sol5 catalyzed a cycloaddition after releasing polyketide chain and the oxidation of side chain. On the other hand, the latter HR-PKS LovB catalyzes a Diels–Alder reaction of linear triene intermediate during the chain extension process (Scheme 1). Before characterization of the monofunctional decalin-forming DAases described in this review, most of polyketide-derived adducts were proposed to be biosynthesized during chain extention stage as in the case of LovB. However, direct and indirect evidence described in this section supported involvement of post-PKS linear precursor in the decalin-forming [4+2] cycloaddition as in the case of Sol5.
Intramolecular DAases generating unique molecular skeletons other than decalin
Spynosyn A (14), a polyketide-derived insecticide isolated from Saccharopolyspora spinosa, possesses a 22-membered macrolactone with a perhydro-as-indacene scaffold.39 Biotransformations of putative synthetic intermediates with several deletion mutants (SpnF/J/L/M) established the biosynthetic machinery for the construction of the tricyclic system.40 In 2011, Liu and co-workers41 revealed that the cyclohexene in the as-indacene core is constructed by SpnF, a homolog of a S-adenosyl-l-methionine (SAM)-dependent methyltransferase, which in fact accelerates the rate of the [4+2] cycloaddition of an α,β,γ,δ-unsaturated macrolactone precursor 15a in 500-fold (Scheme 5). This was the first DAase that catalyzed solely [4+2] cycloaddition different from three bifunctional proteins. The crystal structure of SpnF bound to an S-adenosylhomocysteine (SAH) revealed heavily interaction between SpnF and the co-factor possibly to maintain the structure.42 Subsequent docking simulation suggested predominant hydrophobic contacts with the C5-C6 s-cis state of the macrolactone 15b and a putative hydrogen-bonding interaction between the dienophile and a threonine. Through these interactions, SpnF may facilitate a stabilization of 15b in the preferred transition state to catalyze the Diels–Alder reaction. However, roles of the hydrophobic/hydrogen-bonding interactions remain unclear because of the instability of the cavity-altered mutants. Most recently, quantum mechanical computations and dynamic simulations suggested that the observed [4+2] cycloadduct is constructed by a [6+4] cycloaddition followed by a Cope rearrangement.43
In the recent studies on thiazoyl peptides synthesized ribosomally,44 an interest has been focusing on the formation of a central pyridine ring at the junction of the macrocyclic ring system, which may be biosynthesized by an intramolecular hetero-Diels–Alder reaction of two dehydroalanines and a neighboring carboxyl group (Figure 2). Gene knockout experiments and biochemical analysis of leader peptide modification enzymes facilitated the identification of crucial linear precursor peptides and putative DAases. In 2015, two groups independently reported in vitro functional analysis of the DAases, TclM and TbtD.45, 46 In this reaction, the DAases directly generated the final product without formation of any intermediate was observed. The potential mechanism is proposed in Figure 2; (1) concerted cyclization via a tautomerization of the amide carbonyl group furnishes a tetrahydropyridine; (2) subsequent dehydration followed by an aromatization may proceed spontaneously to form the mature pyridine ring. The promiscuous cyclization activity revealed that chain length and rigidity have no effect on the preorganization of the reactive functionalities in their active site.47 Recently, an unexpected homology of these hetero DAases with elimination enzymes, which afford dehydroalanine/dehydrobutyrine in lantibiotic biosynthesis, was pointed out.48
In addition to decalin-forming DAase PynE3 described in the previous section, spirotetronate/spirotetramate family of antibiotics 8 has a characteristic spiro-conjugate scaffold, constructed by another intramolecular DAase PyrI4 between the diene and the γ-methylene-acyltetronate/tetramate moiety of an acyclic precursor such as 10. Recently, the [4+2] cycloaddition activity of 10 was detected by in vitro reaction with PyrI4, which shows no homology with known enzymes (Scheme 4).32 The homologous VstJ identified in the biosynthetic gene cluster of 12 was also functionally characterized.33 Interestingly, function of PyrI4 was complemented with the homologous enzyme ChlL from the 13 gene cluster,32 suggesting that these enzymes accepted the substrates with various chain lengths and substituent. Presence of homologous DAase genes in this family of the antibiotic gene clusters (VstJ, 26%; KijU, 27%; ChlL, 48% and TcaU4, 26%) will be a suitable signature for searching the structurally related antibiotics.
Recently, crystal structure of monofunctional DAase PyrI4 has been reported (Figure 3).49 Its structure was obtained as PyrI4 alone, and its product bound form in which product 11 was inserted into the active site cavity of rigid β-barrel core and the spirocyclic system of 11 was interacted with hydrophobic portion in the bottom. Hydrogen bonds were observed between cavity residues and tetramate cabonyl and hydroxyl groups, suggesting that those H-bonds act for activation of dienophile in the transition state. Although these interactions were not changed between both states with and without ligand, N-terminal sequence changed the form significantly different in these states, and induced fit interaction between N-terminal sequence and decalin portion/β-barrel core were observed (Figure 3). In this structural change, the unstructured N-terminal sequence formed α-helix which existed as a lid and constrain substrate conformation as transition state. The authors predicted that significant structural change between flexible substrate and rigid product enabled to break interaction between lid-like structure and β-barrel core cavity. This released product from active site, thus avoiding product inhibition. Among nine PyrI4 homologs involving related antibiotic biosynthesis,49 recently characterized DAase VstJ lacked the lid-like N-terminal sequence,33 suggesting that catalytic mechanism is variable in each DAase of this family.
Conclusion and outlook
Basic catalytic mechanism of DAases provided by the antibody DAases50 is very simple; (1) providing a cavity which restricts the conformation to enforce it as transition state; (2) activation of dienophile by hydrogen bonding; (3) escaping from product inhibition. Basically, the essentially same catalytic devices are found in the mechanistic biological study of PyrI4, suggesting these are the only requirement for DAases. Definition of DAase is different to the other enzymes, which are classified by the individual catalytic mechanism and which share common motif and homologous amino acid sequence. However, all of DAases described in this review are distinctive in their catalysis. Considering the fact that ribozymes can also catalyze [4+2] cycloadditions,51 we can recognize the term DAase is rather vague. In addition, if we only rely on the concertedness of the reaction catalyzed, it is very difficult to classify the candidates of putative DAases as Williams and co-workers3 mentioned for the critical discussion on the concertedness in their review; ‘It is also appreciated that tedious, expensive, and time-consuming experiments are required to conduct such a rigorous mechanistic inquiry, a challenge encountered by all workers in this field.′ Reflecting this, none of the candidate DAase currently known has proven its concertedness in its catalysis. Formal DAase can be defined as an enzyme that catalyzes formation of cyclohexene ring from a conjugated diene and an alkene in a stereoselective manner. This may include double Michael reaction, aldol-Michael reaction, biradical reaction and other stepwise reactions. After establishing a detailed reaction mechanism, these members can be classified into several groups. Apart from the definition, cyclohexene-forming enzymes, which are easily modified to accept a variety of substrates, may be useful on the practical point of view.
The bifunctional DAases described in the introduction section have original catalytic activities besides the cycloaddition. On the other hand, the monofunctional DAases SpnF and PyrE3 described in this review have co-factor molecules SAM and FAD, respectively, in their active sites although they do not involve catalysis but they are essential for the cycloaddition. These data suggest that their ancestor enzymes methyltransferase and oxidase provide the cavity for cyclization and then lose their original catalytic activities (Table 1). Due to the simple mechanism of Diels–Alder reaction, activating substrate and stabilizing transition state, any type of enzymes can become DAases. Probably, this is a reason why those bifunctional and monofunctional DAases show no common motif and are phylogenetically distinct from each other.
In the case of four-type DAases Fsa2, PynE3, TclM/TbtD and PyrI4, functionally equivalent enzyme genes were found in the gene clusters of the structurally related metabolites. While the bifunctional enzymes and SpnF do not have functionally related DAases, homologs of the four DAases catalyze similar cycloadditions, suggesting that the latter DAases are likely co-evolved with substrate producer enzymes. Detailed structural and biochemical analysis of PyrI4 showed that PyrI4 and its homolog VstJ did not share common catalytic mechanism. This fact implicates that different mechanism can be operated even if two related DAases catalyze similar cycloadditions. Therefore, comparative study of homologous DAases will provide new aspects to the DAases. In addition, DAases described in this review catalyze construction of complex molecular scaffolds from polyketides and ribosomal peptides. This might be Nature’s strategy for molecular diversification of linear chain intermediates derived from polyketide and ribosomal peptide biosynthetic machinery.
In de novo design of artificial enzyme, which is called a ‘designer enzyme’, researchers choose a suitable scaffold of existing proteins, and then generated computationally designed active site for catalysis of specific Diels–Alder reaction.50, 52 This strategy is very similar to the nature’s strategy generating DAases. Thus, microorganisms likely recruit existing enzymes and then evolutionarily modify them to catalyze a specific [4+2] cycloaddition for creating molecular diversity. The list of natural [4+2] adducts suggests that many natural DAases involve in their biosynthesis. Currently, the designer DAases can handle only simple substrates. On the other hand, most of natural [4+2] adducts have significantly complex and diverse structures. Considering progress in recent 5 years on new natural DAases, elucidation of catalytic mechanism of these DAases as in the case of PyrI4 will provide novel protein scaffolds for DAase and more abundant catalytic mechanisms for entropy trapping and escaping product inhibition. These will help to improve catalytic efficiency of the designer DAases.

Representative examples of the enzymatic decalin formation catalyzed by (a) Sol5 and (b) LovB/LovC.

Reaction mechanism catalyzed by macrophomate synthase.

Decalin formation in the biosynthesis of (a) betaenone and (b) fusarisetin/Sch210972.

Biogenesis of spirotetronate/spirotetramate superfamily of natural products.

Biosynthetic scheme of spinosyn.
References
Nicolaou, K. C., Snyder, S. A., Montagnon, T. & Vassilikogiannakis, G. The Diels–Alder reaction in total synthesis. Angew. Chem. Int. Ed. 41, 1668–1698 (2002).
Juhl, M. & Tanner, D. Recent applications of intramolecular Diels–Alder reactions to natural product synthesis. Chem. Soc. Rev. 38, 2983–2992 (2009).
Klas, K., Tsukamoto, S., Sherman, D. H. & Williams, R. M. J. Org. Chem. 80, 11672–11685 (2015).
Kim, H. J., Ruszczycky, M. W. & Liu, H.-W. Curr. Opin. Chem. Biol. 16, 124–131 (2012).
Oikawa, H. & Tokiwano, T. Enzymatic catalysis of the Diels–Alder reaction in the biosynthesis of natural products. Nat. Prod. Rep. 21, 321–352 (2004).
Oikawa, H. in Comprehensive Natural Products II Chemistry and Biology (eds Mander, L. & Liu, H. -W. 277–314 (Elsevier, Oxford, (2010).
Spole, J., Becker, H., Gupta, M. P., Beith, M. & Huch, V. Novel C-35 terpenoids from the panamanian liverwort Plagiochila moritziana . Tetrahedron 45, 5003–5014 (1989).
Kuo, Y. H., Chan, C. H. & Huang, S. L. A novel bicyclo[2.2.2]octane skeleton diterpene, obtunone, from the heartwood of Chamaecyparis obtusa var. formosana. Chem. Pharm. Bull. 46, 181–183 (1998).
Martins, D., Osshiro, E., Roque, N. F., Marks, V. & Gottlieb, H. E. A sesquiterpene dimer from Xylopia aromatic . Phytochemistry 48, 677–680 (1998).
Hofheinz, W. & Schonholzer, P. Ircinianin, a novel sesterterpene from a marine sponge. Helv. Chim. Acta 60, 1367–1370 (1977).
Sekita, S., Yoshihira, K., Natori, S. & Kuwano, H. Structures of chaetoglobosin A and B, cytotoxic metabolites of Chaetomium globosum . Tetrahedron Lett. 14, 2109–2112 (1973).
Aoyagi, Y. et al. Biomimetic synthesis of grandione from demethylsalvicanol via hetero-Diels–Alder type dimerization and structure revision of grandione. Tetrahedron Lett. 46, 7885–7887 (2005).
Oikawa, H., Katayama, K., Suzuki, Y. & Ichihara, A. Enzymatic activity catalysing exo-selective Diels–Alder reaction in solanapyrone biosynthesis. J. Chem. Soc. Chem. Commun. 1321–1322 (1995).
Kennedy, J. et al. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 284, 1368–1372 (1999).
Watanabe, K. et al. Macrophomate synthase: Characterization, sequence, and expression in Escherichia coli of the novel enzyme catalyzing unusual multistep transformation of 2-pyrones to benzoates. J. Biochem. 127, 467–473 (2000).
Oikawa, H. et al. Macrophomate synthase: Unusual enzyme catalyzing multiple reactions from pyrones to benzoates. Tetrahedron Lett. 40, 6983–6986 (1999).
Fischbach, M. A. & Walsh, C. T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Hoye, T. R. & Dvornikovs, V. Comparative Diels–Alder reactivities within a family of valence bond isomers: a biomimetic total synthesis of (+/-)-UCS1025A. J. Am. Chem. Soc. 128, 2550–2551 (2006).
Ichihara, A., Oikawa, H., Hayashi, K. & Sakamura, S. Structures of betaenones A and B, novel phytotoxins from Phoma betae Fr . J. Am. Chem. Soc. 105, 2907 (1983).
Ichihara, A. et al. A phytotoxin, betaenone C, and its related metabolites of Phoma betae Fr . Agric. Biol. Chem. 47, 2965 (1983).
Ugai, T. et al. Heterologous expression of highly reducing polyketide synthase involved in betaenone biosynthesis. Chem. Commun. 51, 1878–1881 (2015).
Miki, S. et al. Synthesis of (-)-probetaenone I: Structural conformation of biosynthetic precursor of betaenone B. J. Chem. Soc. Perkin Trans. 1, 1228–1229 (1990).
Jang, J. H. et al. Fusarisetin A, an acinar morphogenesis inhibitor from a soil fungus, Fusarium sp. FN080326. J. Am. Chem. Soc. 133, 6865–6867 (2011).
Kato, K. et al. A new enzyme involved in the control of the stereochemistry in the decalin formation during equisetin biosynthesis. Biochem. Biophys. Res. Commun. 460, 210–215 (2015).
Kakule, T. B., Zhang, S., Zhan, J. & Schmidt, E. W. Biosynthesis of the tetramic acids Sch210971 and Sch210972. Org. Lett. 17, 2295–2297 (2015).
Sato, M. et al. Involvement of lipocalin-like CghA in decalin-forming stereoselective intramolecular [4+2] cycloaddition. ChemBioChem 16, 2294–2298 (2015).
Kakule, T. B., Sardar, D., Lin, Z. & Schmidt, E. W. Two related pyrrolidinedione synthetase loci in Fusarium heterosporum ATCC74349 produce divergent metabolites. ACS Chem. Biol. 8, 1549–1557 (2013).
Kakule, T. B. et al. Native promoter strategy for high-yielding synthesis and engineering of fungal secondary metabolites. ACS. Synth. Biol. 4, 625–633 (2015).
Qiao, K., Chooi, Y.-H. & Tang, Y. Identification and engineering of the cytochalasin gene cluster from Aspergillus clavatus NRRL 1. Metab. Eng. 13, 723–732 (2011).
Singh, M. P. et al. Pyrroindomycins, novel antibiotics produced by Streptomyces rugosporus sp. LL-42D005. I. Isolation and structure determination. J. Antibiot. 47, 1250–1257 (1994).
Singh, M. P. et al. Pyrroindomycins, novel antibiotics produced by Streptomyces rugosporus sp. LL-42D005. II. Biological activities. J. Antibiot. 47, 1258–1265 (1994).
Tian, Z. et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265 (2015).
Hashimoto, T. et al. Biosynthesis of versipelostatin: Identification of an enzyme-catalyzed [4+2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J. Am. Chem. Soc. 137, 572–575 (2015).
Jia, X.-Y. et al. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 13, 575–585 (2006).
Zhang, H. et al. Elucidation of the kijanimicin gene cluster: Insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J. Am. Chem. Soc. 129, 14670–14683 (2007).
Fang, J. et al. Cloning and characterization of the tetrocarcin A gene cluster from Micromonospora chalcea NRRL11289 reveals a highly conserved strategy for tetronate biosynthesis in spirotetronate antibiotics. J. Bacteriol. 190, 6014–6025 (2008).
Kasahara, K. et al. Solanapyrone synthase, a possible Diels–Alderase and iterative type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani . ChemBioChem 14, 1245–1252 (2010).
Ma, S. M. et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science 326, 589–592 (2009).
Kirst, H. A. et al. A83543A-D, unique fermentation-derived tetracyclic macrolides. Tetrahedron Lett. 32, 4839–4842 (1991).
Waldron, C et al. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa . Chem. Biol. 8, 487–499 (2001).
Kim, H. J., Ruszczycky, M. W., Choi, S. H. & Liu, H.-W. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 473, 109–112 (2001).
Fage, C. D. et al. The structure of SpnF, a standalone enzyme that catalyzes [4+2] cycloaddition. Nat. Chem. Biol. 11, 256–268 (2015).
Patel, A. et al. Dynamically complex [6+4] and [4+2] cycloadditions in the biosynthesis of spinosyn A. J. Am. Chem. Soc. 138, 3631–3634 (2016).
Walsh, C. T., Acker, M. G. & Bowers, A. A. Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins. J. Biol. Chem. 285, 27525–27531 (2010).
Wever, W. J. et al. Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed forma [4+2] cycloaddition. J. Am. Chem. Soc. 137, 3494–3497 (2015).
Hudson, G. A., Zhang, Z., Tietz, J. I., Mitchell, D. A. & van der Donk, W. A. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J. Am. Chem. Soc. 137, 16012–16015 (2015).
Bowers, A. A., Acker, M. G., Young, T. S. & Walsh, C. T. Generation of thiocillin ring size variants by prepeptide gene replacement and in vivo processing by Bacillus cereus . J. Am. Chem. Soc. 134, 10313–10316 (2012).
Ortega, M. A. et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015).
Zheng, Q. et al. Enzyme-dependent [4+2] cycloaddition depends on lid-like interaction of the N-terminal sequence with the catalytic core in PyrI4. Cell Chem. Biol. 23, 352–360 (2016).
Preiswerk, N. et al. Impact of scaffold rigidity on the design and evolution of an artificial Diels–Alderase. Proc. Natl Acad. Sci. USA 111, 8013–8018 (2014).
Serganov, A. et al. Structural basis for Diels–Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 12, 218–224 (2005).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010).
Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research (A)15H01835 to HO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Minami, A., Oikawa, H. Recent advances of Diels–Alderases involved in natural product biosynthesis. J Antibiot 69, 500–506 (2016). https://doi.org/10.1038/ja.2016.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2016.67
This article is cited by
-
Recent advances in the chemo-biological characterization of decalin natural products and unraveling of the workings of Diels–Alderases
Fungal Biology and Biotechnology (2022)
-
Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels–Alderase
Nature Catalysis (2021)
-
Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold
Nature Chemistry (2021)
-
Discovery and investigation of natural Diels–Alderases
Journal of Natural Medicines (2021)
-
Total synthesis of brevianamide A
Nature Chemistry (2020)