Abstract
Recent years have seen considerable progress in applying single nucleotide polymorphisms (SNPs) to population genetics studies. However, relatively few have attempted to use them to study the genetic differentiation of wild bird populations and none have examined possible differences of exonic and intronic SNPs in these studies. Here, using 144 SNPs, we examined population genetic differentiation in the saker falcon (Falco cherrug) across Eurasia. The position of each SNP was verified using the recently sequenced saker genome with 108 SNPs positioned within the introns of 10 fragments and 36 SNPs in the exons of six genes, comprising MHC, MC1R and four others. In contrast to intronic SNPs, both Bayesian clustering and principal component analyses using exonic SNPs consistently revealed two genetic clusters, within which the least admixed individuals were found in Europe/central Asia and Qinghai (China), respectively. Pairwise D analysis for exonic SNPs showed that the two populations were significantly differentiated and between the two clusters the frequencies of five SNP markers were inferred to be influenced by selection. Central Eurasian populations clustered in as intermediate between the two main groups, consistent with their geographic position. But the westernmost populations of central Europe showed evidence of demographic isolation. Our work highlights the importance of functional exonic SNPs for studying population genetic pattern in a widespread avian species.
Similar content being viewed by others
Introduction
Over recent decades, microsatellite and mitochondrial DNA (mtDNA) have been the two most common molecular markers used in the fields of ecology, evolution and conservation. However, during the last decade, single nucleotide polymorphisms (SNPs) have become increasingly used because of their abundance in the genome, ease of replication in different laboratories and simplicity of analysis (Pariset et al., 2006). Recently, researchers have started to apply SNP-based methods to study the genetic differentiation of wild bird populations, although the studies published so far have mostly used SNPs identified from intron or noncoding sequences (see, for example, Backström et al., 2008; Strand et al., 2012).
In contrast, the potential use of exonic SNPs in revealing population genetic differentiation in species where little demographic isolation is evident using neutral markers has recently been demonstrated to be effective. For example, based on 320 exonic SNPs derived from expressed sequence tags (Moen et al., 2008, Freamo et al. (2011) found that exonic SNPs, especially non-neutral ones, outperformed classical molecular markers (for example, microsatellite and mtDNA) to differentiate the two Atlantic salmon populations with different habitats (rivers and stream vs drainages) and life history characteristics (for example, migration pattern). The accurate population assignment is also an important approach to study immigration and genetic structure (Paetkau et al., 1998), to identify potential admixture of harvested populations (Nielsen et al., 2012) and to analyze the effect of translocation (Witzenberger and Hochkirch, 2008). However, until now, there is no comparison of effectiveness of exonic and intronic SNPs in revealing genetic differentiation of wild populations, although it is well appreciated that exon and intron sequences are subjected to different levels of selection pressure (Lin et al., 2006) and the natural selection can promote genetic differentiation (Lamichhaney et al., 2012). Even worse, the difference between these two types of SNPs are often ignored in recent population genetics studies, including those based on genome (for example, Li et al., 2008; The Bovine HapMap Consortium, 2009) or transcriptome data (for example, Nielsen et al., 2012).
In this study, we aim to test whether introns and exons may give different outputs for the analysis of population genetic differentiation of a widespread avian species. For that aim, we chose the saker falcon as the study species. We sampled birds across Eurasia and obtained the SNP data through amplifying the samples with the EPIC (exon-primed intron crossing; Palumbi and Baker, 1994) primers. Specifically, the EPIC primers were designed on adjacent exon regions and amplified across an intron, which however produced both introns and their flanking exon sequences. In addition, we attempted to explore the possible influence of natural selection on the effectiveness of intronic and exonic SNPs.
Materials and methods
Sample collection, DNA extraction and targeting gene regions
The saker is a raptor inhabiting open landscapes including steppe, desert, agricultural land and plateau grasslands, and has a wide breeding range in Eurasia, extending from central Europe to East Asia (Figure 1a). Two subspecies, exhibiting clinal variation, are generally recognized (Vaurie, 1961; Ferguson-Lees and Christie, 2001) as F. c. cherrug in the west and F. c. milvipes in the east. Recently, Nittinger et al. (2007), using microsatellites and mtDNA, however, revealed little genetic structure in the saker populations they studied and thus did not support subspecies designations. The clinal nature of population differentiation in the saker falcon (Vaurie, 1961; Eastham et al., 2002) presented an ideal system to test the efficiency of intronic and exonic SNPs in the detection of potential population genetic differentiation.
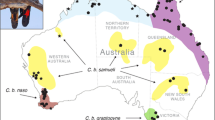
Distribution of saker falcons (a) is based on our surveys and literature (see Supplementary Note for details) and population genetic differentiation reflected by the structure analysis on the intronic SNPs (b, K=2) and exonic SNPs when K=2 (c) and K=3 (d). The 13 sampling sites are labeled and they were grouped as six geographic groups: hollow triangles represent CHQ; hollow circles for DJU; hollow squares for NKZ; pied circles for WKT; black triangles for SEU; and diamond shapes for CEU.
We targeted a sampling to cover most of the Palearctic breeding distribution of the saker (Figure 1a). In total, 136 feather samples comprising 113 plucked contour and 23 molted feathers (mainly flight feathers) were obtained from sites in Croatia, Hungary, Eastern Kazakhstan, Northern Kazakhstan, Western Kazakhstan, Moldova, Mongolia, Qinghai (China), Romania, Slovakia, Turkey, Ukraine and Xinjiang (China; Table 1; Figure 1a). Twenty-five blood samples were collected from live birds and three tissue samples were collected from dead bodies found on or below power lines in Qinghai. Skin samples of historical specimens collected in Bulgaria and Croatia were obtained from the National Museum of Natural History, Sofia (N=7) and the Croatian Natural History Museum, Zagreb (N=2) respectively (Table 1). However, it should be noted that there are regions of the breeding distribution that we were not able to sample, such as southern Central Asia (i.e, Kyrgyzstan, Uzbekistan, Turkmenistan, Tajikistan, Afghanistan and Iran) and Russia, which deserves further efforts in the future.
Plucked feather tips and tissue samples were stored with 99.7% ethanol and molted feathers were kept in dry envelopes. For DNA extraction, feather samples were processed in a fume cupboard in the laboratory of Cardiff University, UK, and DNA extracts were obtained using the Qiagen Blood and Tissue Extraction Kit (Qiagen, Hilden, Germany). We selected only one sample from each nest to minimize the probability of sampling-related individuals. Forty gene fragments from the chicken genome sequence (WASHUC2, www.ensembl.org) were selected, and EPIC primers (Supplementary Table 1) were designed to amplify corresponding sequences from three DNA samples from the east, centre and west of the range (Mongolia, Northern Kazakhstan and Ukraine), respectively. Once found to be polymorphic in these three samples, all individuals were screened using these primers. All sequences were checked against the newly sequenced saker genome (Zhan et al., 2013) using BlastN with E-value=−5. The exon and intron boundaries were defined following the annotated genes in the reference saker genome. In addition, we analyzed exonic variation in two well-characterized gene systems with known exonic variations in vertebrates linked to phenotypic diversity and pathogen resistance, the melanocortin-1-receptor (MC1R; Zhan et al., 2012) and major histocompatibility complex (MHC) Class II B (Alcaide et al., 2008).
Amplification and sequencing
PCR amplifications were performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) in 10 μl containing 1 μl of template DNA, 0.4 μM of the forward and reverse primer, 4 μl of HotStar Taq Master Mix (Qiagen) and 0.2 μg μl−1 bovine serum albumin (New England Biolabs, Hitchin, UK). The amplification of EPIC markers started at 94 °C for 15 min, followed by a touchdown PCR (a total of 45 cycles of 94 °C per 30 s, Tanneal per 30 s, 72 °C min−1) and a final step of 72 °C for 10 min. Tanneal was decreased by 0.5 °C every cycle from 62 °C to the touchdown temperature (52 °C), which was used for following 25 cycles. MC1R amplifications were conducted as described in Zhan et al. (2012). The MHC fragments were amplified using the following profile: 1 cycle of 15 min at 94 °C, 35 cycles of 30 s at 94 °C, 45 s at 56 °C, 1 min at 72 °C and finally, 5 min at 72 °C. We calculated success rates of positive PCRs for each fragment as the number of sequences divided by the total number of feather DNA samples (n=136).
PCR products were sequenced using the original primers and electrophoresed on a 3130XL Genetic Analyzer (Applied Biosystems). To obtain reliable sequences, no less than 10% of total PCR products were randomly selected from each fragment and sequenced from both ends. The final sequences were eye checked and aligned using MEGA (Tamura et al., 2007), from which SNPs were ascertained. Nucleotide diversity (π) was calculated for each identified genetic population (that is, CHQ and ECA in Results) using MEGA.
To verify whether the saker has a single copy of MHC Class II B, we selected all the unique genotypes and cloned the corresponding PCR products using a TOPO TA Cloning Kit (Invitrogen, Paisley, UK) according to the manufacturer’s instructions. Positive colonies were amplified using universal M13 forward and reverse primers (M13F: 5′-GTAAAACGAC-GGCCAG-3′; M13R: 5′-CAGGAAACAGCTATGAC-3′) and 6–8 clones per individual were randomly selected for sequence analysis. Following the standards described in Alcaide et al. (2008), rare cloned sequences found only once and differing by <3 bp from a redundant sequence of the same PCR product were considered as PCR artifacts and assumed to have been sampled already. As recombination of cloned PCR products can result in additional artifacts, comparisons were conducted between cloned sequences and direct sequencing of uncloned PCR products to get reliable polymorphic sites.
Genetic differentiation
Of the 10 EPIC fragments, 9 contained intronic SNPs. Locus S20 was predicted to be a noncoding region in the saker genome but was included in the intronic SNP analysis. Besides MHC and MC1R, four gene fragments (GNB2L1, COE2, CAPZA1 and ANP32B) were found to contain exonic SNPs (Table 2). Arlequin (Excoffier and Lischer, 2010) was used to test linkage disequilibrium between any two SNP loci within each of the two inferred genetic populations (CHQ and ECA in Results). Analysis parameters were set as below: number of permutations=10 000 and number of initial conditions=10. The significant level was set to 0.05, followed by a Bonferroni correction.
For an initial exploration of genetic diversity across Eurasia, we analyzed sakers (Table 1) in six ad hoc geographic groups (Figure 1) mainly according to their geographic proximity and habitat type. The six groups are defined as Chinese Qinghai (comprising the sampling site CHQ on the Tibetan Plateau), Djungaria (DJU; sampling sites MN, CHX and EKZ are grouped into DJU as they represent a continuous population occupying the desert and steppe of the Gobi and Djungaria region), North Kazakhstan (NKZ), West Kazakhstan and Turkey (WKT; TKY and WKZ are grouped as WKT because they are adjacent populations that inhabit arid and saline steppe habitats), Southeast Europe (SEU; including BG, ML, ROM and UKR) and central Europe (CEU; including CR, HUP and SL). We used GenALEX6.5 (Peakall and Smouse, 2012) to calculate observed (HO) and expected heterozygosity (HE), and to test for Hardy–Weinberg equilibrium (HWE) within each group for each locus with α=0.05, followed by a Bonferroni correction. To assess the differentiation between geographic populations, we calculated pairwise D-values (Jost, 2008) using the package DEMEtics (Gerlach et al., 2010) in R. The permissible level of missing data was set to 0.35. Significance levels were determined by 1000 bootstrap resampling, followed by a Bonferroni correction.
Genetic differentiation among ungrouped individuals was first explored using a principal coordinate analysis (PCoA) plot implemented in the software GenALEX. We concatenated the intronic and exonic SNPs into two separate data sets. For exonic SNPs, we also analyzed the MHC and MC1R SNPs separately, because they may be subjected to different selection forces (Alcaide et al., 2008; Bourgeois et al., 2012). Second, we used Bayesian clustering implemented in Structure 2.2 (Pritchard et al., 2000) to examine the genetic partitions within and among saker samples, without reference to their geographic origin. Again, we concatenated the exonic SNPs or intronic SNPs separately. This concatenation can also mitigate against the problem of tight linkage within a single exon or intron sequence (Pritchard et al., 2000). Eight independent runs of K=1–8 were performed with 1 000 000 Markov chain Monte Carlo repetitions after 50 000 burn-in period. The missing alleles were included in the analysis. The admixture ancestry model was chosen and allele frequencies were assumed to be correlated. The LOCPRIOR model (Hubisz et al., 2009) was used and the locations set according to PopData. K was identified using the maximal values of ΔK based on the rate of change in the log probability of data between successive K values (Evanno et al., 2005). Graphical output was displayed using Distruct v1.1 (http://rosenberglab.bioinformatics.med.umich.edu/distruct.html).
For the two genetic clusters identified by structure (see Results), D-values were again obtained with DEMEtics, and HO, HE and HWE examined using GenALEX. Furthermore, within each cluster, mean HE for MHC SNPs was compared with those of intronic or other exon SNPs using a Kolmogorov–Smirnov test implemented in PAST (Hammer et al., 2001).
Selection analysis
We analyzed whether the SNP allele frequencies could have been modified by selection by dividing the samples into the two genetic clusters revealed in the structure analysis. All the exonic and intronic SNPs were used for the analysis. For the three nests (two in CHQ and one in ROM) where exonic and intronic genotypes were obtained from different feather samples, only intronic SNPs were included in the analysis. As a result, the total of 147 samples with 140 typed SNP loci were used to identify selected SNPs. The first selection test uses a nonhierarchical island population model to detect FST outlier loci and was employed using ARLEQUIN 3.5. As we considered only two groups without substructure, this test is equivalent to the algorithm developed by Beaumont and Nichols (1996). The permissible level of missing data was set to 0.1 and 200 000 coalescent simulations were performed. After completing the analysis, we performed false discovery rate correction on P-values using QVALUE (Storey 2002) with lambda=0. We considered loci with P<0.01 and q-value<0.05 as candidate non-neutral SNPs.
The second test uses a Bayesian estimation of the coancestry coefficient FST to decide whether a signature of selection can be inferred for a particular locus (BayeScan; Foll and Gaggiotti, 2008). We used the program defaults of 20 pilot runs of 5000 iterations and an additional burn-in of 50 000 iterations, then 1 000 000 iterations with a sample size of 50 000 and thinning interval of 20. For candidate-selected SNPs, the false discovery rate was also set to <0.05.
Previous studies based on simulated data have indicated that isolation by distance (IBD) can lead to false positives in tests of population genetic structure and the detection of loci under selection (Meirmans, 2012). We investigated IBD using a Mantel test implemented in GenALEX. Linearized FST values between any two sampling sites (Table 1) were checked for correlation with (log-transformed) geographic distance. The P-values were calculated using 9999 permutations. The Bulgarian (BG) and Croatian (CR) samples were excluded from this analysis because, except for one sample, the coordinates of the museum samples were unavailable. There is only one Slovakian sample in the exonic SNP data set, which was also removed from the analysis.
Results
Genome position, PCR efficiency and MHC verification
From the 40 EPIC markers designed from the chicken genome sequences, we obtained 10 saker PCR fragments located on 10 different chicken chromosomes (Table 2). When compared with the saker genome (Zhan et al., 2013), the 12 fragments (the 10 EPIC fragments plus MHC and MC1R) could be mapped onto 11 genome scaffolds (NCBI: AKMU01000000) and 11 annotated genes (Table 2). MHC and RPL13 were found to be on the same scaffold but there was no exonic SNP identified in RPL13. Thus, all fragments analyzed could be inferred to have been generated from independent genome regions (Table 2). When taking into account their positions on the saker falcon genome (Zhan et al., 2013), the average physical distance among introns and exons is about 1.5 and 0.6 Mb respectively, although both distances are very likely to be underestimated because we only have scaffold information for the saker genome. As expected, the linkage disequilibrium analysis suggested that, within saker populations (CHQ and ECA, respectively), the linkage disequilibrium generally existed among SNPs from the same fragment, but no consistent linkage disequilibrium signal was detected among the markers from different fragments.
The PCR efficiency of saker feather (plucked+molted feather) DNA extracts varied substantially: LDH-B had the lowest positive PCR rate (36.8% in Table 2), but the success rate reached 74.3% for the MHC. In general, fragment length was found to be significantly negatively correlated with PCR efficiency (Pearson’s r=−0.78; P=0.01). From the 12 fragments, a total of 5761 bp intron and 1491 bp exon sequences were obtained. Overall, 108 SNPs were detected within the saker intron sequences, ∼1 per 53 bp.
MHC cloning (23 individuals) consistently showed only one or two alleles for each individual using the criteria above, suggesting that the MHC Class II B gene is present as a single copy in the saker. When checked against the saker genome, no stop codons or frameshift mutations were observed in the sequenced MHC exons, implying they are not pseudogenes. Twelve SNPs were found in an alignment of only 269 bp MHC, which confirmed the observation that the second exon of MHC gene is one of the most highly polymorphic in vertebrates (see also Alcaide et al., 2008). Seventeen SNPs were identified from the length of 601 bp in MC1R and one non-synonymous SNP mutation occurred exclusively in three sakers of the population CHQ.
Genetic diversity partition among six geographic populations
Observed and expected heterozygosity values within each of the six geographic samples are shown in Table 3. For the intronic SNP, both HO (0.073–0.111) and HE (0.092–0.122) were similar across the populations. No significant departures from HWE were detected in the populations from CHQ, NKZ and WKT, but seven, eight and three of 108 SNPs deviated from HWE in DJU, SEU and CEU, respectively (Supplementary Table 2). For the exonic SNPs, although all the HO values were similar among the populations, HE in CHQ (0.156) was higher than those in the five other populations. Of the 36 exonic SNPs, 6 deviated from HWE in SEU, but no more than 3 SNPs were detected in any other population (details in Supplementary Table 2).
Pairwise population comparisons based on exonic SNPs showed that the genetic differentiation (D) between CHQ and the other populations was always one order of magnitude larger, and was significantly differentiated (Table 4). However, of the five Europe/central Asia comparisons (ECA, including DJU, NKZ, WKT, SEU and CEU), only two pairwise populations (CEU and DJU; SEU and DJU) were found to be significantly differentiated. In the analysis of intronic SNP, although still significant, the pairwise D-values between CHQ and the other populations were intermediate (Table 4).
Population differentiation and selection between eastern and western sakers
No strong pattern of partitioning among samples across the two principle components could be detected using the 108 intronic SNP loci (Figure 2a), although it should be noted that the majority of sampled populations grouped together in axis 1 while separating into two clusters in axis 2. The concatenated exonic SNPs (n=36; Figure 2b) showed a clearer pattern than intronic SNPs in that axis 1 separated more populations (11/14) into two clusters. Extensive within-population partitioning suggests that both concatenation systems failed to detect systematic among-population genetic differentiation in sakers. Interestingly, the PCoAs of the MC1R separated some individuals from the CHQ population (in red in Figure 2c) from other sakers. In addition, the PCoA results of MHC were more partitioned, with most sakers from Qinghai (CHQ) being separated from other samples (in red in Figure 2d) and the Kazakhstan (EKZ, NKZ and WKZ), Xinjiang (CHX; China) and most Mongolia samples positioned intermediate between CHQ and the more western populations (circled areas in Figure 2d).
Structure analysis largely supported the PCoA results. Although the intronic SNPs failed to uncover any population differentiation (Figure 1b), the analysis of exonic SNPs demonstrated a strong signal of differentiation between the CHQ and other saker populations (Figure 1c) in that the ΔK reached its maximum value when K=2 (Supplementary Table 3). Further, Structure analysis revealed genetic differentiation within Europe/central Asia (ECA) to some extent when K=3 was analyzed (Figure 1d): the two genetic backgrounds being found with one mainly restricted to central Europe (Slovakia, Croatia and Hungary) and Turkey. However, it should be noted that the sample sizes of Romania, Croatia and Turkey populations are small (LamichhaneyTable 1) and the PCR efficiency for the Slovakian samples was lowest. When K=3, evidence for an eastern genetic background (that is, CHQ) was also uncovered in some Kazakhstan (NKZ, EKZ and WKZ), Xinjiang (CHX), Mongolia (MN), Croatia (CR) sakers and, interestingly, museum samples from Bulgaria and Croatia (Figure 1d). It is also noted that, using the same exonic SNP data, the structure analysis without lociprior information showed similar pattern of population genetics of sakers (CHQ vs others; Supplementary Figure 1). However, the simulations contained more noise (Supplementary Figure 1a vs Figure 1c). Furthermore, the ΔK reached its maximum when K=4 but it could not provide more meaningful information than K=2 in that CHQ again has a relatively distinct background but each of the remaining populations had fairly admixed ancestry from multiple genetic clusters (Supplementary Figure 1c). These differences confirmed the conclusion that the LOCPRIOR model is necessary for inferring population structure when the signal of structure is weak (for example, Pritchard et al., 2000).
For the saker populations in ECA, mean intronic HO and HE values were 0.096 and 0.116, whereas they were 0.048 and 0.079 for the exonic SNPs (Supplementary Table 2). Five out of 36 exonic and 18 of 108 intronic SNPs were found to deviate from HWE (Supplementary Table 2). For the MHC, no significant difference was found between the HE values and those of other markers with all the P-values more than 0.05 in the Kolmogorov–Smirnov tests. The average nucleotide diversity of intron sequences was 0.0073 (0–0.0460) for CHQ and 0.0082 (0.0018–0.0219) for the ECA population (Table 2). Between the CHQ and ECA populations, exonic D was 0.055 and the two populations were found to be significantly genetically different (P=0.001). The intronic analysis suggested a smaller D-value (0.012).
Between the CHQ and ECA, Arlequin simulations indicated there were five SNPs under directional selection (Figure 3, q<0.05) with three exonic SNPs in the MHC gene and one intronic SNP for each of S18 and S29 (Figure 3). In contrast, the BayeScan analysis could not find any selection signal (q<0.05). Pairwise estimates of population differentiation showed no significant IBD inferred from either exonic (R2=0.0287, P=0.12) or intronic SNPs (R2=0.0027, P=0.33; Supplementary Figures 2a and b).
Discussion
Feathers as a source of DNA for SNP-based studies of wild birds
Feather samples are one of the most common DNA sources for ornithological studies (Katzner et al., 2012) and have previously been used for amplifying mitochondrial sequences and nuclear microsatellites in a phylogeographic study of saker falcons (20% of total sample size in Nittinger et al., 2007). In our study, about 79% of DNA extracts were from feather samples (n=136) and we found that, as expected, shorter fragments yielded significantly higher PCR efficiencies (Table 2). This is not unexpected because feathers usually contain less DNA (see, for example, Bush et al., 2005) that is sometimes more degraded than in blood and tissue samples (Harvey et al., 2006). Another factor affecting the PCR efficiency is storage condition. In our experiment, most failed PCRs came from a single site (Slovakia), where storage ethanol had evaporated during transport prior to analysis. However, for elusive species inhabiting vast and remote areas, traditional sampling methods (for example, blood or tissue) can be prohibitively expensive. SNP technology can potentially be applied to very small PCR fragments and therefore provide the most viable method for population genomic analysis of noninvasively collected samples.
Population genetic diversity
The intronic SNP rate (one per 53 bp) found in the saker populations is similar to that in Larus dominicanus (one per 79 bp; Dantas et al., 2009) but considerably higher than those in Ficedula albicollis (one per 345 bp; Backström et al., 2008) and Thryothorus pleurostictus (one per 305 bp; Crammer et al., 2008). It is also noted that the saker intronic SNP rate here is much higher than the frequency of heterozygous SNP sites (one per 1282 bp) inferred in the saker genome analysis (Zhan et al., 2013). This discrepancy should be partly due to the absence of homozygous SNPs in the genome study (n=1), but moreover because the current estimate is based on more than 100 sakers from different populations (Table 1).
Furthermore, the average nucleotide diversity (π=0.0073 or 0.0082) inferred from intronic sequences is within the same range with those found in L. dominicanus (0.00151<π<0.00599; Dantas et al., 2009), F. albicollis (0.0029; Backström et al., 2008) or Parus caeruleus (0.0018; Backström et al., 2008). Our results therefore indicate that the saker retains considerable genetic diversity, although its populations have been greatly reduced in parts of its range.
Contrasting population genetic differentiation revealed by intronic and exonic SNPs
The observed intrapopulation partitioning (Figures 2a and b) shows that both PCoAs of concatenated SNP data sets did not uncover interpopulation genetic differentiation in sakers. One explanation for this observation is that allele frequencies for different SNP loci have diverged differently within and among populations, and as a consequence, the distance-based PCoA could not distinguish these populations because of complex selection signatures existing in the whole concatenated intronic or exonic SNPs. Furthermore, intronic SNPs lacked power to differentiate any populations (Figures 1b and 2a; Supplementary Table 3), except that the differentiation index (D) showed genetic differentiation to some extent (Table 4). In contrast, exonic markers demonstrated their efficiency in partitioning saker populations. First, both Bayesian clustering and genetic distance-based PCoA of either MHC or MC1R suggested that the Qinghai population (CHQ) is differentiated from the others, a result strongly supported by the results of genetic differentiation estimates among the six a priori-grouped geographic populations with the largest D-values and most significant differentiation found between CHQ and the other populations (Table 4). However, there were several individuals assigned to different genetic clusters within CHQ (Figure 1b), possibly the result of selection or very recent gene flow.
Second, within the Europe/central Asia populations, even exonic SNPs could not differentiate sakers from DJU (MN, CHX and EKZ,), NKZ, WKT (WKZ only) and SEU (UKR, ML, ROM and BG) populations (Figure 1d), suggesting substantial gene flow among saker populations in Europe/central Asia. Alternatively, the current exonic SNP system does not have sufficient resolving power for separating saker populations in this region. In that case, more strongly selected genes or genome-level data are needed to fully resolve their genetic differentiation, as has recently been demonstrated in fish species that have low neutral estimates of genetic differentiation (for example, Freamo et al., 2011; Nielsen et al., 2012). Third, the exonic SNP-based analysis showed that within ECA, the central Europe and Turkey populations were differentiated from others to some extent (Figure 1d). It is known that the central European population suffered drastic declines in the 1960s and 1970s (Baumgart, 1991) and declines have also been documented in Turkey (Dixon et al., 2009). Along with drift, historical population bottlenecks might have driven differentiation among saker populations in this region. On the basis of similar sample coverage in the Europe and central Asia, Nittinger et al. (2007) found that most sakers in central Europe possessed the same mtDNA haplogroup, consistent with our findings. However, different from the ‘distinct’ CEU population we infer, their ‘CEU’ mitochondrial haplogroup also occurred in northern Kazakhstan (Nittinger et al., 2007), suggesting that different evolutionary processes might occur for mtDNA and nuclear exons of northern Kazakhstan sakers. Finally, three historical museum samples from Bulgaria (N=2) and Croatia (N=1) exhibited an eastern genetic background (Figure 1d) indicating genetic exchange across an unfragmented, contiguous historical population, or alternatively, the unusual genetic background is a consequence of genetic drift.
Both Nittinger et al. (2007) and our exonic SNP analysis do not support the subspecific division of F. c. cherrug and F. c. milvipes in sakers (Ferguson-Lees and Christie, 2001). However, compared with classical mtDNA genes and nuclear microsatellites (Nittinger et al., 2007), our exonic SNPs were found to be better to uncover the genetic differentiation of current saker populations, although the low level of differentiation of microsatellite markers might be an artifact from homoplasy and high variation within populations (Alcaide et al., 2008). It is noted that Nittinger et al.’s (2007) work did not include samples from significant parts of the sakers’ distribution range such as the Tibetan Plateau. Furthermore, the use of FST in their work (Nittinger et al., 2007) might also account for this discrepancy, as FST statistic can be very sensitive to certain parameters such as the number of alleles in the markers or the differences in population sizes (Jost, 2008).
Forces shaping saker population genetic differentiation
It is known that drift, selection and migration are the three major forces shaping the genetic structure in moderately sized populations (Smith, 1994). However, in the saker, genetic drift is expected to have limited influence on current population differentiation primarily because each of the main samples (CHQ and ECA) was estimated to originate from a large and geographically widespread population (Dixon, 2009). Migration may have a role in shaping this pattern but it should counteract differentiation (Smith, 1994). Therefore, it seems that selection is a good candidate to explain the differentiation of CHQ and ECA. Our FST-based analysis provides support for this hypothesis and showed that three genes RPL13, COE2 and MHC exhibit signatures of directional selection. This result also explained why MHC was more robust in differentiating saker populations than MC1R gene (Figure 2c vs Figure 2d).
As shown in Supplementary Figure 3, the common alleles of the selected SNPs in the ECA population show reduced diversity compared with those in CHQ. In addition, they also lack heterozygosity (for example, HE in Supplementary Table 2). When signals of this kind are detected, the SNPs may have been subjected to selection sweeps and have become nearly fixed, as a consequence in the ECA saker populations (Pickrell et al., 2009). Alternatively, the relatively distinct nature of CHQ could be because of the higher altitude and specific habitat of the region (the Tibetan Plateau), which might exert different selection pressures on sakers there. For example, local adaptation has been reported for the plateau human populations. Simonson et al. (2010) concluded that positively selected haplotypes of EGLN1 and PPARA were thought to be responsible for high-altitude adaptation, whereas Yi et al. (2010) found the strongest signal of natural selection on one SNP at EPAS1, which is the downstream target of EGLN1 in the hypoxia-inducible factor pathway (Simonson et al., 2010). However, given vast spatial distribution of both the CHQ population and the other genetic cluster found in the data, the environment is likely to be highly heterogeneous, precluding meaningful environmental associations in our work.
In contrast with Arlequin, BayeScan analysis did not uncover any selection signatures using the same data set. This may be because BayeScan is more conservative in identifying selected SNPs, as reported by Zhao et al. (2013), or because there remain false positives in the Arlequin FST results even with false discovery rate correction. The scarcity of selected SNP can result from gene flow between ECA and CHQ (Figures 1c and d), which made the FST values underestimated. The selection analysis may also suffer few and short gene fragments examined (Table 2), which however are typical for noninvasive genetic studies. IBD is thought to be another potential problem for inferring the population genetic structure and detecting the selected SNPs (Meirmans, 2012). However, our Mantel analysis found no significant correlation between the pairwise genetic differentiation and geographic distance in the studied saker populations regardless of SNP types (Supplementary Figure 2). In another study using microsatellites, Nittinger et al. (2007) drew the same conclusion. Therefore, it seems that ‘IBD’ should have a limited influence on the population genetic differentiation of saker falcons.
Interestingly, we found three MHC SNPs under directional selection but none under balancing selection (Figure 3). This is at odds with previous reports where the main mechanism for MHC evolution is thought to be balancing selection (Alcaide et al., 2008; Landry and Bernatchez, 2001). However, if balancing selection is the reason for shaping genetic structure of saker falcons, we would expect an excess of heterozygotes in the saker populations due to the heterozygote advantage (Landry and Bernatchez, 2001). On the contrary, all the MHC SNPs were in HWE within the two genetic clusters of saker falcons (Supplementary Table 2) and the HE values we observed at the MHC were not significantly different from those of other markers. These results are also distinct from the lesser kestrel (F. naumanni), for which Alcaide et al. (2008) identified balancing selection as the main mechanism for determining the MHC-based population genetic structure. A possible explanation is that lesser kestrels occupy more homogeneous habitats covering mid-latitude and low altitudes of Eurasia, whereas saker falcons not only share a similar range but live on the Tibetan Plateau. These two distinct ecoregions could facilitate directional selection against certain MHC alleles, conferring greater susceptibility to local pathogens.
In summary, our results suggest different roles of intronic and exonic SNPs in understanding population biological questions. Whereas analysis of intronic SNPs indicates a nearly homogeneous background of the current saker falcons, exonic SNP analyses uncovered at least two genetic clusters from the current populations where the Qinghai population has separated from the rest. We also identified Central Eurasian populations as conduits for genetic exchange between the eastern and western saker populations, although we cannot rule out the possibility that their mixed background was due to the low discriminatory power of current exon system. Ancient or occasional recent hybridization has been reported between saker falcons and other falcon species (especially gyrfalcon F. rusticolus; Nittinger et al., 2005, 2007; Johnson et al., 2007), which may also influence the population history of saker falcons. The efficiency of our SNP system in the examination of this interspecific hybridization warrants future research.
Data Archiving
Sequence data have been deposited at GenBank (JX543974-JX544062; JX666614-JX667684).
References
Alcaide M, Edwards SV, Negro JJ, Serrano D, Tella JL . (2008). Extensive polymorphism and geographical variation at a positively selected MHC class II B gene of the lesser kestrel (Falco naumanni). Mol Ecol 17: 2652–2665.
Backström N, Fagerberg S, Ellegren H . (2008). Genomics of natural bird populations: a gene-based set of reference markers evenly spread across the avian genome. Mol Ecol 17: 964–980.
Baumgart W . (1991) Der Sakerfalke. Neue Brehm Bücherei, Bd 514. Ziemsen Verlag: Wittenberg.
Bourgeois YXC, Bertrand JAM, Thébaud C, Milá B . (2012). Investigating the role of the melanocortin-1 receptor gene in an extreme case of microgeographical variation in the pattern of melanin-based plumage pigmentation. PLoS One 7: e50906.
Beaumont MA, Nichols RA . (1996). Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263: 1619–1626.
Bush KL, Vinsky MD, Aldridge CL, Paszkowski CA . (2005). A comparison of sample types varying in invasiveness for use in DNA sex determination in an endangered population of greater Sage-Grouse (Centrocercus uropihasianus). Conserv Genet 6: 867–870.
Cramer ERA, Stenzler L, Talaba AL, Makarewich CA, Vehrencamp SL, Lovette IJ . (2008). Isolation and characterization of SNP variation at 90 anonymous loci in the banded wren (Thryothorus pleurostictus). Conserv Genet 9: 1657–1660.
Dantas GP, Godinho R, Morgante JS, Ferrand N . (2009). Development of new nuclear markers and characterization of single nucleotide polymorphisms in kelp gull (Larus dominicanus). Mol Ecol Resour 9: 1159–1161.
Dixon A . (2009). Saker falcon breeding population estimates. Part 2. Asia Falco 33: 4–10.
Dixon A, Ragyov D, Ayas Z, Deli MT, Demerdzhiev D, Angelov I et al. (2009). Population status of breeding saker falcons (Falco cherrug) in Turkey. Avian Biol Res 2: 213–220.
Eastham CP, Nicholls MK, Fox NC . (2002). Morphological variation of the saker (Falco cherrug) and the implications for conservation. Biodivers Conserv 11: 305–325.
Evanno G, Regnaut S, Goudet J . (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Lischer HEL . (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567.
Ferguson-Lees J, Christie DA . (2001) Raptors of the World. Christopher Helm: London, UK.
Foll M, Gaggiotti O . (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993.
Freamo H, O'Reilly P, Berg PR, Lien S, Boulding EG . (2011). Outlier SNPs show more genetic structure between two Bay of Fundy metapopulations of Atlantic salmon than do neutral SNPs. Mol Ecol Resour 11 (Suppl 1): 254–267.
Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P . (2010). Calculations of population differentiation based on Gst and D: forget Gst but not all of statistics!. Mol Ecol 19: 3845–3852.
Hammer Ø, Harper DAT, Ryan PD . (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4: 9.
Harvey MG, Bonter DN, Stenzler LM, Lovette IJ . (2006). A comparison of plucked feathers versus blood samples as DNA sources for molecular sexing. J Field Ornithol 77: 136–140.
Hubisz M, Falush D, Stephens M, Pritchard J . (2009). Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332.
Johnson JA, Burnham KK, Burnham WA, Mindell DP . (2007). Genetic structure among continental and island populations of gyrfalcons. Mol Ecol 16: 3145–3160.
Jost L . (2008). Gst and its relatives do not measure differentiation. Mol Ecol 17: 4015–4026.
Katzner TE, Wheeler M, Negro JJ, Kapetanakos Y, DeWoody JA, Horvath M et al. (2012). To pluck or not to pluck: scientific methodologies should be carefully chosen, not ‘one size fits all’. J Avian Biol 43: 15–17.
Landry C, Bernatchez L . (2001). Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar). Mol Ecol 10: 2525–2539.
Lamichhaney S, Barrioa AM, Rafatia N, Sundströma G, Rubina C-J, Gilberta ER et al. (2012). Population-scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proc Natl Acad Sci USA 109: 19345–19350.
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S et al. (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104.
Lin H, Zhu W, Silva JC, Gu X, Buell CR . (2006). Intron gain and loss in segmentally duplicated genes in rice. Genome Biol 7: R41.
Meirmans PG . (2012). The trouble with isolation by distance. Mol Ecol 21: 2839–2846.
Moen T, Hayes B, Baranski M, Berg PR, Kjøglum S, Koop BF et al. (2008). A linkage map of the Atlantic salmon (Salmo salar) based on EST-derived SNP markers. BMC Genomics 9: 223.
Nielsen EE, Cariani A, Aoidh EM, Maes GE, Milano I, Ogden R et al. (2012). Gene-associated markers provide tools for tackling illegal fishing and false eco-certification. Nat Commun 3: 851.
Nittinger F, Haring E, Pinsker W, Wink M, Gamauf A . (2005). Out of Africa? phylogenetic relationships between Falco biarmicus and the other Hierofalcons (Aves: Falconidae). J Zool Syst Evol Res 43: 321–331.
Nittinger F, Gamauf A, Pinsker W, Wink M, Haring E . (2007). Phylogeography and population structure of the saker falcon (Falco cherrug) and the influence of hybridization: mitochondrial and microsatellite data. Mol Ecol 16: 1497–1517.
Paetkau D, Shields GF, Strobeck C . (1998). Gene flow between insular, coastal and interior populations of brown bears in Alaska. Mol Ecol 7: 1283–1292.
Palumbi SR, Baker CS . (1994). Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol Biol Evol 11: 426–435.
Pariset L, Cappuccio I, Marsan PA, Dunner S, Luikart G, England PR et al. (2006). Assessment of population structure by single nucleotide polymorphisms (SNPs) in goat breeds. J Chromatogr B Analyt Technol Biomed Life Sci 833: 117–120.
Peakall R, Smouse PE . (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539.
Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D et al. (2009). Signals of recent positive selection in a worldwide sample of human populations. Genome Res 19: 826–837.
Pritchard JK, Stephens M, Donnely P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Simonson TS, Yang YZ, Huff CD, Yun HX, Qin G, Witherspoon DJ et al. (2010). Genetic evidence for high-altitude adaptation in Tibet. Science 329: 72–75.
Smith PJ . (1994). Genetic diversity of marine fisheries resources: possible impacts of fishing. FAO Fishery. FAO: Rome, Italy. Technical Paper No. 344, p 57.
Storey JD . (2002). A direct approach to false discovery rates. J R Statist Soc Ser B 64: 479–498.
Strand TM, Segelbacher G, Quintela M, Xiao L, Axelsson T, Höglund J . (2012). Can balancing selection on MHC loci counteract genetic drift in small fragmented populations of black grouse? Ecol Evol 2: 341–353.
Tamura K, Dudley J, Nei M, Kumar S . (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
The Bovine HapMap Consortium. (2009). Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324: 528–532.
Witzenberger KA, Hochkirch A . (2008). Genetic consequences of animal translocations: a case study using the field cricket, Gryllus campestris L. Biol Conserv 141: 3059–3068.
Vaurie C . (1961). Systematic notes on palearctic birds, no. 45, Falconidae: the genus Falco (part 2). Am Mus Novit 2038: 1–24.
Yi X, Liang Y, Huerta-Sanchez E, Jin X, Zha XPC, Pool JE et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78.
Zhan XJ, Dixon A, Fox NC, Bruford MW . (2012). Missense SNP of the MC1R gene is associated with plumage variation in the Gyrfalcon (Falco rusticolus). Anim Genet 43: 460–462.
Zhan XJ, Pan SK, Wang JY, Dixon A, He J, Muller MG et al. (2013). Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyle. Nat Genet 45: 563–566.
Zhao SC, Zheng PP, Dong SS, Zhan XJ, Wu Q, Guo XS et al. (2013). Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet 45: 67–71.
Acknowledgements
This work was funded by the Environment Agency-Abu Dhabi (EAD) and we thank HE Mohammed Al Bowardi for his interest and support. XZ was partially supported by the Recruitment Program of Global Youth Experts of China. We also thank the following saker biologists for kindly providing samples and making this study possible: Andrey Munteanu (ML), Mehmet Deli (TKY), Jóska Fidloczky (HUP), and Zlatozar Boev (BG); Pan Shengkai, He Jing and Dong Shanshan for assisting with data analysis; Samrat Mondol for SNP isolation; and Nick Fox for discussion on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Zhan, X., Dixon, A., Batbayar, N. et al. Exonic versus intronic SNPs: contrasting roles in revealing the population genetic differentiation of a widespread bird species. Heredity 114, 1–9 (2015). https://doi.org/10.1038/hdy.2014.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.59
This article is cited by
-
Common patterns in the molecular phylogeography of western palearctic birds: a comprehensive review
Journal of Ornithology (2021)
-
Analysis of pooled genome sequences from Djallonke and Sahelian sheep of Ghana reveals co-localisation of regions of reduced heterozygosity with candidate genes for disease resistance and adaptation to a tropical environment
BMC Genomics (2019)
-
Genome-wide SNP analyses reveal high gene flow and signatures of local adaptation among the scalloped spiny lobster (Panulirus homarus) along the Omani coastline
BMC Genomics (2018)
-
The first set of universal nuclear protein-coding loci markers for avian phylogenetic and population genetic studies
Scientific Reports (2018)
-
A candidate gene-based association study reveals SNPs significantly associated with bud burst in European beech (Fagus sylvatica L.)
Tree Genetics & Genomes (2015)