Abstract
Within the interleukin-10 receptor 1 (IL10R1) gene, two common variants are associated with certain diseases: single-nucleotide polymorphism 3 (SNP3), a serine-138 to glycine mutation is in linkage disequilibrium with SNP4, a glycine-330 to arginine mutation, both of which are considered loss-of-function alleles. However, the molecular consequence of G330R is unknown. We investigated possible roles of G330R on the dynamics of IL10R1 surface expression and signal transducer and activator of transduction (STAT) phosphorylation. HeLa cells expressing the respective IL10R1 haplotype were stimulated with IL-10. Significant reduction of IL10R1 surface expression was observed after ligand binding. Receptor expression remained low on continuous incubation with IL-10. In contrast, when treated with an IL-10 pulse, IL10R1 surface expression returned to its resting state within 3–9 h irrespective of the haplotype. STAT3 was rapidly phosphorylated both in cells with wild-type (WT) or variant IL10R1, and maintained phosphorylated when cells were cultured with IL-10. On IL-10 pulse, however, STAT3 phosphorylation declined rapidly in cells expressing IL10R1-G330R but not IL10R1-WT or S138G. Similar dynamics were observed with STAT1 phosphorylation at Tyr701. No differences in janus kinase 1 (JAK1) activation were observed in cells with WT or variant IL10R1. Our results indicate that IL10R1-G330R does not alter surface expression but duration of STAT phosphorylation, indicating that the position of G330 is important in stabilizing the STAT signal.
Similar content being viewed by others
Introduction
Genetic variation contributes to susceptibility or resistance of disease. This is also true for genes in the interleukin-10 (IL-10) signaling pathway. Polymorphisms within IL-10 pathway genes contribute to chronic hepatitis C, tuberculosis, human immunodeficiency virus, systemic lupus, rheumatoid arthritis and the outcome of organ or bone marrow transplantation.1 Most recently, IL-10 receptor mutations have been associated with severe early-onset enterocolitis,2 a phenotype that had been observed with IL-10-knockout mice some 16 years earlier.3
IL-10 is a pleiotropic cytokine that acts on many hematopoietic cells. It was first described as a cytokine synthesis inhibitory factor secreted from CD4+ T-cells to terminate the inflammatory responses.4 Thus, the principal function of IL-10 appears to be the termination of inflammatory responses. IL-10 inhibits the production of cytokines (for example, tumor necrosis factor-α, IL-1, IL-6, IL-8 and interferon-γ) and chemokines (CC and CXC); it regulates the proliferation and differentiation of T-cells, B-cells, antigen-presenting cells, natural killer cells, mast cells and granulocytes.5 The ability of cells to respond to IL-10 depends on the expression of the IL-10 receptor complex, which is composed of two subunits: IL-10 receptor 1 (IL10R1) and IL10R2. Stimulation of this receptor complex leads to the activation of janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2) and phosphorylation of signal transducer and activator of transcription (STAT) 1, 3 (and in some cells also STAT5 (ref. 6)), which translocate to the nucleus and induce gene expression. Both receptor subunits belong to the class II cytokine receptor superfamily and function as tetramers.5, 7 Although IL10R1 has a dominant role in ligand binding and signal transduction, IL10R2 participates in the initiation and transduction of the signal.8 In contrast to IL10R1, which is expressed mainly by cells of the immune system,9 IL10R2 is ubiquitously expressed and serves as a second subunit for other receptor complexes.10 Therefore, distinct binding domains of IL10R1 are assumed to be responsible for the IL-10 specificity.5
Two common variants within the IL10R1 gene were associated with schizophrenia,11 liver fibrosis in chronic hepatitis C12 and various autoimmune diseases.13 Substitutions of glycine-330 to arginine (G330R, herein termed SNP4) and of serine-138 to glycine (S138G, SNP3) are known to affect the sensitivity of the cell to respond to IL-10 through incompletely understood mechanisms.14 The extracellular SNP3 has been demonstrated to interfere with ligand binding.15, 16 However, the mechanistic consequences of the intracytoplasmic SNP4 are unclear. After ligand binding, murine IL10R1 undergoes rapid internalization and proteosomal degradation, a process which is described to be dependent on certain intracytoplasmic residues (that is, murine amino acids 282–389).17 As SNP4 is located in this very region, we primarily tested whether this variant alters the dynamics of IL10R1 expression in response to IL-10. As the same IL10R1 region is also sensitive for interaction with JAK1 (amino acids 300–578 interact with the JH7–6 domain of JAK1 (refs 18, 19)), we also tested for the dynamics of IL10R1/JAK1 binding as well as JAK1, STAT1 and STAT3 phosphorylation as an alternative hypothesis.
Results
Reduction of IL10R1 surface expression after ligand binding
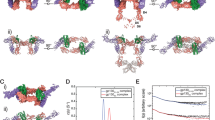
IL10R1 is thought to internalize after ligand binding and, thus, decrease at the cell surface. To test for the timing of such event, HeLa cells expressing wild-type (WT) IL10R1 (IL10R1-WT) were stimulated with 50 ng ml−1 rhIL-10. Receptor expression was measured at 5, 10, 20, 30, 60 and 120 min of IL-10 stimulation. IL-10 binding leads to a fast reduction of IL10R1 cell surface expression (Figures 1a and b). IL10R1 expression changed within 5 min. Stable reduction was achieved within 30 min of IL-10 treatment (Figure 1b), a time point that was chosen for further studies.
Decline of IL10R1 expression on IL-10. HeLa cells transfected with IL10R1-WT were treated with 50 μg ml−1 rhIL-10. After 0, 5, 10, 20, 30, 60 and 120 min of incubation, IL10R1 expression was analyzed by flow cytometry. (a) Histograms of FL2 show cells stained with anti-IL10R1-phycoerythrin antibody (empty) and cells stained with an isotype control (gray). (b) IL10R1 expression (mean fluorescence intensity) of four independent experiments (each performed in triplicates) was normalized to non-treated control and is presented as the fold change in MFI for each time point.
IL10R1 surface expression returns to resting state after IL-10 pulse
As IL10R1 expression is lowered on ligand binding, we next determined whether SNP4 alters such dynamics. SNP4 naturally occurs either alone or together with SNP3 (so called SNP3+4).14 Thus, the effect of both haplotypes was investigated. HeLa cell clones stably transfected with pIRESpuro3-IL10R1-WT, -IL10R1-SNP4 or -IL10R1-SNP3+4 were treated with IL-10 for 30 min. In one set of the experiment, IL-10 was removed (referred to as ‘IL-10 pulse’). In another set, IL-10 was left in the medium for the entire length of the experiment (referred to as ‘IL-10 cont.’). As control, HeLa cells were grown without IL-10 (referred to as ‘w/o IL-10’). The cells were incubated for 0.5, 3, 6, 9, 12 or 24 h, and IL10R1 expression was analyzed by flow cytometry (Figure 2). Cells transfected with variant IL10R1 showed similar basal expression levels of the receptor as compared with the WT receptor (Figure 2b). A significant reduction of IL10R1 cell surface expression was observed both in WT and variant IL10R1 readily after 30 min of IL-10 treatment (Figure 2c). IL10R1 surface expression returned to normal levels within 3–9 h after removal of IL-10. When cells were continuously cultured with IL-10, the receptor expression remained low. Treatment conditions (IL-10 pulse versus IL-10 cont.) had a significant influence on the IL10R1 cell surface expression (P<0.001), independent of the SNP status. No difference was observed between the IL10R1-WT, IL10R1-SNP4 or IL10R1-SNP3+4, concerning the dynamics of receptor expression (P=0.762). Pulse treatment revealed a significant relation between post-pulse culture period and the level of IL10R1 (P<0.005), indicating that IL10R1 returned to the cell surface in a time-dependent manner.
SNP4 does not affect the dynamics of IL10R1 expression on ligand binding. (a) Schematic diagram of the experimental procedure. w/o IL-10: cells were grown without IL-10. IL-10 pulse: cells were treated with IL-10 (50 ng ml−1) for 30 min, and further incubated without IL-10. IL-10 cont.: cells were kept in the presence of IL-10. IL10R1 cell surface expression was measured at indicated time points. (b) Basal receptor levels of HeLa cells transfected with IL10R1-WT, -SNP4 or -SNP3+4 were compared using flow cytometry. Histogram plots of FL2 show overlays of cells stained with anti-IL10R1-phycoerythrin antibody (empty) and cells stained with an isotype control (gray). (c) HeLa cells transfected with IL10R1-WT, -SNP4 or -SNP3+4 were cultured with or without IL-10 or treated with an IL-10 pulse. IL10R1 expression was measured by flow cytometry at indicated time points. Data represent the mean±s.e.m. of nine (IL10R1-WT), six (IL10R1-SNP4) or four (IL10R1-SNP3+4) independent experiments. Data are fold change in MFI for each time point compared with control.
Alteration of IL10R1 expression during cell cycle
Although a cytomegalovirus promoter should provide constitutive IL10R1 expression by stable transfected HeLa cells, some variation of IL10R1 expression was observed within each time point. Several studies demonstrated a relationship between cell cycle distribution and expression of surface proteins, including the interferon,20 estrogen21 and progesterone receptors.22 Thus, we further tested IL10R1-WT clones after synchronization with nocodazole, which arrested almost all cells in the G2/M phase of the cell cycle (Figure 3a). Removal of nocodazole allowed the cells to progress through the cell cycle (Figure 3b). Furthermore, it has been reported that IL-10 mediates proliferation in several cell types.5 In our experiments, addition of IL-10 had no influence on cell cycle progression. However, the more cells were in G0/G1 the higher was the IL10R1 mean fluorescence intensity (MFI; Figure 3c top; r=0.534, P=0.037). Inversely, the more cells in G2/M the lower was the IL10R1 MFI (Figure 3c bottom; r=0.443, P=0.075), indicating that IL10R1 density decreases with cell cycle progression from G1 to G2/M phase. Experiments with IL10R1-SNP4 cells showed the same change from G1 to G2/M phase (data not shown). It is likely that the alterations of IL10R1 expression during cell cycle contributed to intra- and inter-experimental variation in receptor expression.
Alteration of IL10R1 expression throughout cell cycle. (a) HeLa cells transfected with IL10R1-WT were treated with nocodazole for 14 h and analyzed for DNA content on staining with propidium iodide (+noc). Non-treated cells served as a control (−noc). (b) HeLa cells transfected with IL10R1-WT were arrested in G2/M by nocodazol and further cultured with or without IL-10 for 0, 3, 6, 9, 12 and 24 h. The percentage of cells within the G0/G1, S and G2/M phase is shown. Data represent mean±s.d. of three independent experiments. (c) Correlation between IL10R1 expression levels and percentage of cells within G0/G1 (r=0.534; P=0.037), S (r=0.253; P=0.214) or G2/M phase (r=−0.443; P=0.075) are shown. MFI of IL10R1 expression are presented as the fold change for each time point compared with t=0. Correlations (r) and significance (P) were calculated by Pearson's correlation analysis.
The intracytoplasmic IL10R1-SNP4 allows a rapid reduction in STAT phosphorylation
To investigate possible changes of JAK–STAT activation dynamics, we analyzed IL-10-induced STAT3 phosphorylation at Tyr705 and dephosphorylation in cells expressing IL10R1-WT, -SNP3, -SNP4 or the combined SNP3+4 receptor haplotype. Similar to the experiments above, cells were treated with IL-10 pulse (in order to examine STAT3 dephosphorylation dynamics) and compared with continuous IL-10 cultivation. After 0.5, 3, 6, 9, 12 or 24 h, STAT3 activation was analyzed from whole-cell extracts by phospho-specific western blotting. STAT3 was phosphorylated within 30 min in response to IL-10 (Figure 4a). When cultured with IL-10 (Figure 4 ‘IL-10 cont.’), no differences were seen between the various IL10R1 haplotypes, indicating that none of the receptor variants influenced the speed of STAT3 phosphorylation. However, when IL-10 was removed from culture after 30 min (Figure 4 ‘IL-10 pulse’), a fast decrease of STAT3 phosphorylation was observed as early as of 3 h in both SNP4 carrying haplotypes. SNP3 (S138G), within the IL10R1 gene, was described as a loss-of-function allele based on its reduced IL-10-induced STAT1 and STAT3 phosphorylation.15 The duration of STAT activation was similar to cells expressing the WT receptor. IL10R1-WT or IL10R1-SNP3 haplotypes displayed slow STAT3 dephosphorylation (the activation lasted over 24 h). The same kinetics were observed when protein extracts were blotted for STAT1 phosphorylation at Tyr701 (figure 4b). The variant arginine-330 leads to a timely reduced period of the receptor's STAT activation, suggesting a role of glycine-330 in stabilizing STAT phosphorylation.
Dynamics of STAT3, STAT1 and JAK1 phosphorylation in HeLa cells expressing WT or mutant IL10R1. HeLa cells transfected with IL10R1-WT, -SNP3, -SNP4 or -SNP3+4 were cultured in the absence (−) or presence of IL-10 (IL-10 cont.), or treated with an IL-10 pulse for 30 min (IL-10 pulse). After 0.5, 3, 6, 9, 12 and 24 h, cells were lysed and phosphorylation of STAT3 at Tyr705 (a) and STAT1 at Tyr701 (b) was detected by western blot. (c) JAK1 phosphorylation was detected after 6 h (6 h IL-10 or 30 min IL-10 pulse).
We also performed immunoprecipitation experiments to verify binding of JAK1 to IL10R1 in WT and SNP4 or SNP3+4 containing haplotypes. JAK1 binding remained unaffected on continuous IL-10 treatment (6 h) or pulse treatment (6 h, 30 min IL-10; data not shown). We further analyzed activation of JAK1 using pJAK1 antibody. Western blot analysis showed that JAK1 phosphorylation (Tyr 1022/1023) was unaltered in all the cell lines investigated (Figure 4c).
Discussion
Several reports have demonstrated relation between abnormalities of IL-10 signaling and susceptibility or resistance to disease.12, 15, 23, 24, 25 Severe IL-10 receptor mutations have been associated with early-onset inflammatory bowel diseases,2 thereby proving the importance of this receptor in maintenance of intestinal immune homeostasis. Cell-type-specific expression of IL10R1 is assumed to be a critical factor for the ability of cells to respond to IL-10. This may explain the pleiotropic outcome of IL-10-mediated signaling in vivo.13 Changes of the IL10R1 cell surface expression have impressive consequences for IL-10 bioactivity.26, 27, 28 However, the surface expression pattern and its functional consequences of IL10R1 are incompletely understood. The hypothesis of this study was that IL10R1-G330R modulates IL10R1 expression or STAT activation on ligand stimulation, and thereby interferes with IL10R1 signaling. Our results demonstrate that IL10R1-G330R does not alter the dynamics of receptor expression. However, G330R caused a marked reduction in the duration of STAT1 and STAT3 phosphorylation after IL-10 pulse treatment, indicating that the position of G330 is important in stabilizing the STAT signal. Our findings are in line with earlier studies, demonstrating a loss-of-function for IL10R1-G330R.14
G330 is located in one of the two regions on the IL10R1, which are necessary for the interaction with JAK1 (amino acids 300–578 interacts with the JH7-6 domain on JAK1), the so-called Box 2B motif.18, 19 Mutations in JAK1-binding regions of certain cytokine receptors, including interferon-γRα, have been shown to decrease the interaction with JAK1 and/or impair JAK1 activation.18 In further experiments, we investigated possible effects of G330R on destabilizing JAK1 binding to IL10R1, thereby affecting the persistence of STAT phosphorylation and allow premature dephosphorylation. However, we did not find any differences in JAK1 binding or its activation on IL-10 treatment. Tyrosine phosphorylation of JAK1 remained constant up to 6 h with WT and variant receptor. These results indicate that JAK1 activation is not affected by the exchange of glycine-330 to arginine. Consistent with findings of others,29 our results showed IL-10 induced downstream signaling (STAT phosphorylation) up to 24 h with WT-IL10R1. In contrast, STAT phosphorylation on ligand stimulation was not sustained in SNP4, as was observed with pulse treatment of IL-10. This suggests the possibility of ligand binding having an effect on the sustainability of STAT signal in this IL10R1 haplotype. It has been demonstrated that mutations in IL-10 receptors result in impairment of IL-10 signaling in inflammatory bowel diseases.2 Our observation that WT-IL10R1 can sustain the STAT activation in the absence of ligand, but not IL10R1-G330R, might have implications. As STAT3 activity is important for suppressing inflammation, decrease in IL-10 concentration in the inflamed tissue might aggravate disease in the presence of IL10R1-G330R with impairment in the duration of STAT3 phopsphorylation.
Murine IL10R1 undergoes rapid internalization after IL-10 binding followed by proteasome-mediated degradation.17 Our studies with human IL10R1 confirm reduction of the receptor expression on ligand binding. Although the amount of IL-10 used (50 ng ml−1) should be sufficient to bind all receptor molecules on the cell surface, a small amount of IL10R1 (even though reduced) remained present throughout the time-course. This can be explained by the fact that the expression of both receptor subunits, IL10R1 and IL10R2, is necessary for many cells to initiate signal transduction in response to IL-10. It is likely that IL10R1 is overexpressed with our constructs. Thus, the amount of IL10R1 molecules with all four constructs we used exceeds the amount of IL10R2 molecules on the cell surface. This was shown in a previous study by our group.30 It seems unlikely that our findings can be explained by sterical hindrance of the receptor detection (by the phycoerythrin-labeled antibody) through IL-10. In fact when IL-10 was removed after a stimulatory pulse, IL10R1 expression was delayed by up to 9 h. If IL-10 had obstructed the binding of the antibody, IL10R1 detection should have been restored readily after IL-10 removal. Therefore, the reduction of cell surface receptor on IL-10 represents likely a true reduction of receptor on the cell surface through internalization.
It is important to understand whether the ligand-induced receptor internalization is needed for the diversity of IL10R1 signals.31 At this point, we have to reject our hypothesis that SNP4 changes IL10R1 expression dynamics, but rather suggest the involvement of proteins negatively regulating STAT activation. In studies with murine IL10R1 variants, a region between amino acids 282–389 was described to be involved in signal termination.17 Deletion of this domain led to enhanced IL-10 signaling due to deficient receptor degradation. It was hypothesized that this region may interact with phosphatases or other inhibitors of the IL10R1 signaling pathway. In contrast to receptor truncations in which the whole region was missing, here we show that a single exchange of glycine to arginine in this region resulted in the opposite effect: decreased IL-10 signaling in terms of reduced duration of STAT phosphorylation. It is possible that G330R induces negatively regulatory proteins, such as phosphatases, by providing better interaction sites. In addition, the possibility that in the absence of ligand, docking of STAT on IL10R1 is altered in this SNP cannot be ruled out. Further experiments are required to elaborate the mechanism behind sustainability of STAT phosphorylation in IL10R1-G330R, including the role of phosphatases.
Though we were working with single-cell clones of IL10R1-transfected HeLa cells, IL10R1 expression varied between different time points and experiments. Our data suggest that IL10R1 expression is low in G2/M phase, increases in G1 phase, and remains relatively constant during the S phase. This might be caused by an increase in cell size before the cell division. Our methodology is uncertain about the absolute number of IL10R1 on the surface. Last, IL10R1 induced STAT phosphorylation but did not change the progression of the cells through the cell cycle, suggesting that IL-10 has no growth effects on HeLa.
In summary, our studies revealed a reduction of human IL10R1 cell surface expression on ligand binding and on progression through the cell cycle, both of which are independent of the G330R variant. Also, G330R did not affect JAK1 binding to IL10R1 or activation. However, this variant caused a rapid reduction of STAT3 and also STAT1 phosphorylation, thereby reducing the duration of the intracellular IL-10 signal, a finding that may explain the loss-of-function of the G330 allele.14
Materials and methods
Cells and cell culture
HeLa cells stably transfected with pIRESpuro3-IL10R1-WT (clone #21, 31, 38, 39, 41), -SNP3 (clone #28, 30), -SNP4 (clone #10, 11, 21, 23, 43) or -SNP3+4 (clone #7, 12, 23, 26) were maintained in RPMI supplemented with 10% fetal calf serum and 1 μg ml−1 puromycine (Sigma-Aldrich, St Louis, MO, USA).15 Cells expressing either the WT or variant IL10R1 were seeded in six-well plates at a density of 2.5 × 105 cells per well. For stimulation, cells were treated with 50 ng ml−1 rhIL-10 (Bender MedSystems GmbH, Vienna, Austria) for the indicated length of time or with IL-10 pulse by which IL-10 was removed after 30 min by washing the cells twice with phosphate-buffered saline. Cells were collected after further incubation in RPMI containing 10% fetal bovine serum for different time periods. In some experiments, cells were synchronized with nocodazole (100 μg ml−1 in dimethyl sulfoxide; Sigma-Aldrich) for 14 h.
Flow cytometry
After appropriate stimulation, IL10R1 cell surface expression was analyzed as described.15 Phycoerythrin-conjugated anti-hIL10R1 antibody (Pharmingen, San Diego, CA, USA) or phycoerythrin-conjugated isotype control (IgG2a, κ; BD Biosciences, Mountain View, CA, USA) were used. For cell cycle analysis, cells were washed twice with ice-cold phosphate-buffered saline and fixed in 100% ice-cold ethanol for up to 24 h at 4 °C. Cells were then centrifugated for 5 min at 1500 r.p.m. and stained with propidium iodide solution (20 mg ml−1 propidium iodide, 60 mg ml−1 RNAse in phosphate-buffered saline) at room temperature in the dark for 10 min. Cell cycle distribution was analyzed by flow cytometry using CellQuest acquisition software (FACScan; BD Biosciences).
Western blot analysis
Immunoblotting was performed as previously described.15 Phospho-STAT3 (Tyr 705), phospho-STAT1 (Tyr701), phospho-JAK1 (Tyr1022/1023), STAT3 and STAT1 monoclonal antibodies (all from Cell Signaling Technology, Danvers, MA, USA) were used. Detection was carried out with horseradish peroxidase-conjugated secondary antibodies (horseradish peroxidase-conjugated anti-rabbit immunoglobulin G, GE Healthcare, Little Chalfont, BKM, UK; horseradish peroxidase-conjugated anti-mouse immunoglobulin G, Abcam, Cambridge, MA, USA). For pJAK1, infrared-dye-conjugated secondary antibody was used and detected with Odyssey infrared imaging (LI-COR Biotechnology, Lincoln, NE, USA).
Statistics
Flow cytometry data were analyzed descriptively. Differences in the IL10R1 expression concerning the experimental condition (IL-10 continuous or pulse treatment) and SNP composition were analyzed for statistical significance using a linear mixed effects model. Data are presented as the decimal logarithm of the fold change in MFI for each time point compared with the negative control±s.e.m. The log MFI fold change was defined as an independent variable and time point, SNP composition and experimental condition as fixed effects. Correlation between cell cycle and IL10R1 expression was estimated from flow cytometry data using linear regression analysis. The amount of cells within each cell cycle phase was compared with the median MFI of IL10R1 cell surface expression measured at the same time point. P-values below 0.05 were considered to be statistically significant.
References
Asadullah K, Sterry W, Volk HD . Interleukin-10 therapy--review of a new approach. Pharmacol Rev 2003; 55: 241–269.
Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009; 361: 2033–2045.
Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W . Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993; 75: 263–274.
Fiorentino DF, Bond MW, Mosmann TR . Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 1989; 170: 2081–2095.
Moore KW, de Waal MR, Coffman RL, O’Garra A . Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 2001; 19: 683–765.
Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM . IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett 1996; 394: 365–370.
Josephson K, Logsdon NJ, Walter MR . Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 2001; 15: 35–46.
Lutfalla G, Gardiner K, Uze G . A new member of the cytokine receptor gene family maps on chromosome 21 at less than 35 kb from IFNAR. Genomics 1993; 16: 366–373.
Liu Y, Wei SH, Ho AS, de Waal MR, Moore KW . Expression cloning and characterization of a human IL-10 receptor. J Immunol 1994; 152: 1821–1829.
Commins S, Steinke JW, Borish L . The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol 2008; 121: 1108–1111.
Schosser A, Aschauer HN, Wildenauer DB, Schwab SG, Albus M, Maier W et al. Homozygosity of the interleukin-10 receptor 1 G330R allele is associated with schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 347–350.
Hofer H, Neufeld JB, Oesterreicher C, Grundtner P, Wrba F, Gangl A et al. Bi-allelic presence of the interleukin-10 receptor 1 G330R allele is associated with cirrhosis in chronic HCV-1 infection. Genes Immun 2005; 6: 242–247.
Tilg H, van Montfrans C, van den EA, Kaser A, Van Deventer SJ, Schreiber S et al. Treatment of Crohn's disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut 2002; 50: 191–195.
Gasche C, Grundtner P, Zwirn P, Reinisch W, Shaw SH, Zdanov A et al. Novel variants of the IL-10 receptor 1 affect inhibition of monocyte TNF-alpha production. J Immunol 2003; 170: 5578–5582.
Grundtner P, Gruber S, Murray SS, Vermeire S, Rutgeerts P, Decker T et al. The IL-10R1 S138G loss-of-function allele and ulcerative colitis. Genes Immun 2009; 10: 84–92.
Gruber SG, Gloria LM, Grundtner P, Zdanov A, Gasche C . Differential signaling of cmvIL-10 through common variants of the IL-10 receptor 1. Eur J Immunol 2008; 38: 3365–3375.
Wei SH, Ming-Lum A, Liu Y, Wallach D, Ong CJ, Chung SW et al. Proteasome-mediated proteolysis of the interleukin-10 receptor is important for signal downregulation. J Interferon Cytokine Res 2006; 26: 281–290.
Usacheva A, Sandoval R, Domanski P, Kotenko SV, Nelms K, Goldsmith MA et al. Contribution of the Box 1 and Box 2 motifs of cytokine receptors to Jak1 association and activation. J Biol Chem 2002; 277: 48220–48226.
Usacheva A, Kotenko S, Witte MM, Colamonici OR . Two distinct domains within the N-terminal region of Janus kinase 1 interact with cytokine receptors. J Immunol 2002; 169: 1302–1308.
Takane H, Ohdo S, Yamada T, Koyanagi S, Yukawa E, Higuchi S . Relationship between diurnal rhythm of cell cycle and interferon receptor expression in implanted-tumor cells. Life Sci 2001; 68: 1449–1455.
Ikegami A, Inoue S, Hosoi T, Kaneki M, Mizuno Y, Akedo Y et al. Cell cycle-dependent expression of estrogen receptor and effect of estrogen on proliferation of synchronized human osteoblast-like osteosarcoma cells. Endocrinology 1994; 135: 782–789.
Cassanelli S, Guillaud P, Louis J, Seigneurin D . Image cytometry of progesterone receptor expression during the cell cycle in the MCF-7 cell line. J Histochem Cytochem 1991; 39: 1713–1718.
Asadullah K, Eskdale J, Wiese A, Gallagher G, Friedrich M, Sterry W . Interleukin-10 promoter polymorphism in psoriasis. J Invest Dermatol 2001; 116: 975–978.
Eskdale J, McNicholl J, Wordsworth P, Jonas B, Huizinga T, Field M et al. Interleukin-10 microsatellite polymorphisms and IL-10 locus alleles in rheumatoid arthritis susceptibility. Lancet 1998; 352: 1282–1283.
Hermann J, Gruber S, Neufeld JB, Grundtner P, Graninger M, Graninger WB et al. IL10R1 loss-of-function alleles in rheumatoid arthritis and systemic lupus erythematosus. Clin Exp Rheumatol 2009; 27: 603–608.
Crepaldi L, Gasperini S, Lapinet JA, Calzetti F, Pinardi C, Liu Y et al. Up-regulation of IL-10R1 expression is required to render human neutrophils fully responsive to IL-10. J Immunol 2001; 167: 2312–2322.
Tamassia N, Calzetti F, Menestrina N, Rossato M, Bazzoni F, Gottin L et al. Circulating neutrophils of septic patients constitutively express IL-10R1 and are promptly responsive to IL-10. Int Immunol 2008; 20: 535–541.
Ding Y, Qin L, Zamarin D, Kotenko SV, Pestka S, Moore KW et al. Differential IL-10R1 expression plays a critical role in IL-10-mediated immune regulation. J Immunol 2001; 167: 6884–6892.
Williams L, Bradley L, Smith A, Foxwell B . Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J Immunol 2004; 172: 567–576.
Gruber SG, Gloria LM, Grundtner P, Zdanov A, Gasche C . Differential signaling of cmvIL-10 through common variants of the IL-10 receptor 1. Eur J Immunol 2008; 38: 3365–3375.
Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H et al. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 2008; 321: 663–668.
Acknowledgements
This study was supported by generous funds of the Austrian Science Fund (P17943 to CG). We thank Paul Grundtner for establishing the HeLa cell clones, Chukwuma Agu, Sabine Gruber and Gloria Luciani for technical advice, Thomas Decker (Max F Perutz Laboratories, University of Vienna) for providing materials.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Finsterbusch, M., Khare, V., Campregher, C. et al. An intracytoplasmic IL-10 receptor variant permits rapid reduction in STAT3 activation. Genes Immun 12, 575–581 (2011). https://doi.org/10.1038/gene.2011.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2011.12
Keywords
This article is cited by
-
Interleukin-10 receptor-1 expression in monocyte-derived antigen-presenting cell populations: dendritic cells partially escape from IL-10's inhibitory mechanisms
Genes & Immunity (2015)
-
Exome sequencing identifies novel compound heterozygous mutations of IL-10 receptor 1 in neonatal-onset Crohn's disease
Genes & Immunity (2012)