Abstract
Intellectual disability (ID) is a clinically complex and heterogeneous disorder, which has variable severity and may be associated with additional dysmorphic, metabolic, neuromuscular or psychiatric features. Although many coding variants have been implicated in ID, identification of pathogenic non-coding regulatory variants has only been achieved in a few cases to date. We identified a duplication of a guanine on chromosome X, NC_000023.10:g.69665044dupG 7 nucleotides upstream of the translational start site in the 5ʹ untranslated region (UTR) of the known ID gene DLG3 that encodes synapse-associated protein 102 (SAP102). The dupG variant segregated with affected status in a large multigenerational family with non-syndromic X-linked ID and was predicted to disrupt folding of the mRNA. When tested on blood cells from the affected individuals, DLG3 mRNA levels were not altered, however, DLG3/SAP102 protein levels were. We also showed by dual luciferase reporter assay that the dupG variant interfered with translation. All currently known pathogenic DLG3 variants are predicted to be null, however the dupG variant likely leads to only a modest reduction of SAP102 levels accounting for the milder phenotype seen in this family.
Similar content being viewed by others
Introduction
Intellectual disabilities (ID) are frequent, highly heterogeneous disorders primarily characterised by limitations in intellectual functioning, memory and adaptive behaviour and diagnosed before the age of 18.1 The study of X-chromosome-linked forms of ID has lead the way in the discovery of pathogenic gene variants and molecular pathways implicated in ID, forming the foundations for current systematic large-scale variant discovery efforts.2, 3 Improved phenotyping and unbiased genetic screens using whole-exome or -genome sequencing (WES or WGS) of larger cohorts of affected individuals together with powerful computational tools has markedly accelerated ID gene discovery.2, 3 However, these studies have been most successful in the discovery of coding or gene-disrupting variants, whereas the identification of non-coding and regulatory variants has rarely been attempted.4 In addition, the current success is biased to severe syndromic forms of ID, whereas the larger part, that of mild to moderate non-syndromic ID, is considerably less well understood. Over the past two decades we have invested considerable effort to study large X-chromosome-linked families that have undergone extensive genetic testing and remained unresolved.4 In one such large family of >140 individuals (family 312)5 we used WGS to identify a single guanine duplication in the 5ʹ untranslated region (UTR) of the DLG3 mRNA that segregates with the affected status. We show that although the extra guanine does not affect DLG3 mRNA levels, it perturbs the efficiency of DLG3 mRNA translation, leading to reduction of DLG3 protein, which is the most likely cause of ID in this family.
Materials and methods
Detailed description of materials and methods can be found in Supplementary Information.
Results
Clinical findings
Clinical information (n=10) is presented in Supplementary Table 1. The affected male individuals showed mild to moderate ID, no dysmorphism, below average height and head circumference and did not have any specific health concerns. A minority of female carriers reportedly had mild learning difficulties.
Gene identification
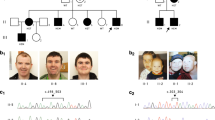
We predicted X-linked recessive inheritance of ID in the family based on the segregation of the disease phenotype (Figure 1a). Linkage and haplotype analysis through 16 family members using markers specific for chromosome X (chrX) (Supplementary Tables 2 and 3) revealed two significant linkage peaks with a maximum LOD score of 2.40 and 2.05 (at θ =0.01), respectively (Supplementary Figure 1). The first linkage peak mapped to Xq13 between markers DXS1125 and DXS559 (hg19 chrX:68289376_70881390) (Supplementary Table 3). The second linkage peak mapped to Xq23 between markers DXS1059 and DXS8067 (hg19 chrX:111325969_119360393). The disease-associated haplotypes were present in all affected males (where DNA was available for testing) except for the monozygotic twins (VI-12 and VI-13).
(a) Multi-generation pedigree. The pedigree represents a classical X-linked recessive inheritance model with mostly affected males and carrier females. The affected individuals indicated by shaded boxes or circles, mildly affected individuals by semi-shaded shapes and the carriers are denoted by a dot in the circle. The solid outer circle indicates the proband, whereas the dotted outer circles indicate individuals that were subjected to Sanger sequencing. (b) A stretch of G nucleotides within DLG3 5ʹ UTR is conserved among mammalian species. Alignment of 5ʹ UTR sequences immediately upstream of DLG3 mRNA translational initiation start site is shown. A set of six conserved G nucleotides in the wild-type sequence is boxed.
Using multiple different screening strategies, we covered all open reading frames in the linkage intervals however no coding variants that passed filtering criteria and segregated with ID were identified (Supplementary Tables 4 and 5). From a single affected male (IV-1; Individual LOVD ID 58821) WGS we identified 167 747 variants of any type on chrX. No copy number, large structural variants or transposable element insertions fulfilled our candidate prioritisation criteria (Supplementary Table 4). There were 33 non-coding variations located within the linkage interval, of which only eight were associated with a known ID gene (Supplementary Table 6). Only the DLG3 5ʹ UTR (NC_000023.10:g.69665044dupG) variant was predicted to have a regulatory effect (RegulomeDB score of 3a). The variant is located at a moderately conserved site (Figure 1b, average GERP score for the polyG tract 3.15±1.12) only seven base pairs from the ATG start codon. Sanger sequencing (where DNA was available) confirmed segregation of the guanine duplication in all affected individuals and carriers with the disease haplotype identified by linkage (Supplementary Figure 2).
Dual luciferase reporter assay and RNA structure prediction
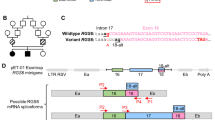
To determine the impact of the NC_000023.10:g.69665044dupG variant, we cloned the full length (−347) or partial (−119) mutant or control 5ʹ UTR and the first nine nucleotides of the open reading frame of DLG3 mRNA (retaining the Kozak sequence) upstream of and in-frame with the firefly luciferase coding sequence under the control of a thymidine kinase promoter (Figure 2a). The relative firefly luciferase activity was significantly reduced in HEK293T cells transfected with the reporter plasmid carrying the full-length mutant 5ʹ UTR compared with the control sequence and this effect occurred in a dose-dependent manner (Figure 2a). No reduction was observed for the shorter mutant 5ʹ UTR reporter (Figure 2a). The reduced reporter gene activity was not a result of reduced transcription because no significant decline in the firefly luciferase mRNA levels between both short and full-length mutant or control 5ʹ UTR reporter transfected HEK293T cells was observed (Supplementary Figures 3a and b). We also showed that DLG3 mRNA levels were similar, albeit moderately higher in the affected LCLs compared with a set of control LCLs (Figure 2d and Supplementary Figure 3c). However, synapse-associated protein 102 (SAP102) levels were reduced in the affected compared with control LCLs (Figures 2b and c). We therefore argued that the decreased firefly luciferase activity could be due to altered structure of the mutant 5ʹ UTR sequence affecting the efficiency of translation and translation initiation in particular. Indeed, in silico prediction showed folding of the full-length (but not the short) DLG3 5ʹ UTR was significantly disrupted by the G duplication compared with the wild type (Supplementary Figure 4). Three other 5ʹ UTR variants present in the ExAC database had no effect on the RNA structure (Supplementary Figure 4).
Duplication of a G nucleotide within DLG3 5ʹ UTR interferes with translation. (a) HEK293T cells were transfected with different amounts of reporter constructs and pRL-TK transfection control plasmid. Luciferase activity was normalised to Renilla luciferase. The experiment was repeated three times where each treatment was done in triplicate and assayed three times. The values mean±SD is shown. Significance was calculated by Student’s t-test. (b) Levels of SAP102 protein in LCLs from affected and control individuals. The levels of SAP102 and β-tubulin loading control were measured by quantifying the band intensities using ImageJ software. SAP102 levels relative to β-tubulin control are shown. (c) Mean±SD SAP102/β-tubulin calculated from values in b. Significance was tested by non-parametric Mann–Whitney U-test. (d) Mean±SD DLG3 mRNA levels calculated from Supplementary Figure 3c.
Discussion
The relative ease of undertaking WGS coupled with substantially reducing costs of sequencing has now opened unprecedented avenues for identifying both coding and noncoding disease variants (eg, regulatory, microRNA, lncRNA, 5ʹ and 3ʹ UTR variants) that previously would have escaped identification. These technical developments coincide with recent findings linking regulatory variants in XLID. For example, with targeted enrichment and sequencing of both coding and noncoding regions located within the linkage interval of the MRX3 family we found a variant in the HCFC1 promoter region. This variant abolished binding of YY1 transcription factor to its cognate DNA-binding sequence and resulted in upregulation of HCFC1 transcription, which is the likely cause of non-syndromic ID in the MRX3 family.4 Detection of differential gene expression in patient-derived cells with an X-chromosome cDNA microarray was used to implicate a promoter variant that altered the binding of ELK1 transcription factor and thus reduced PLP2 expression in XLID.6
DLG3 is a known XLID gene that encodes SAP102 that is a member of the membrane-associated guanylate kinase (MAGUK) family of proteins. MAGUKs are a group of ionotropic scaffolding proteins located at the post-synaptic density (PSD) including PSD-95, PSD-93, PSD-97 and SAP102 that are involved in formation and plasticity of excitatory synaptic terminals of neurons in the brain.7 SAP102 possesses three tandem PDZ (PSD-95/Disc large/Zona occludens) domains at the amino-terminus, middle SH3 (Src Homology 3) domain and an inactive carboxy-terminal GK (Guanylate kinase) domain.5 In rat, SAP102 is highly expressed in dendrites and axons in the foetal and early-late post-natal hippocampus and is reduced at 6 months, whereas PSD-95 and PSD-93 expression increase with age.8 DLG3 was the first ID gene to be linked to NMDA receptor-mediated signalling and synaptic plasticity.5 Dlg3 knockout mice show cognitive deficits with a specific spatial learning deficit that is attenuated by extended training.8 Furthermore, SAP102 depletion increases the number of elongated filopodia, which is commonly seen in mouse models and ID patients.9 That SAP102 has a critical role in early brain development is consistent with reports showing that loss-of-function variants are associated with XLID.5, 10, 11, 12 PSD-95 and SAP102 have both overlapping and specific functions.8 Less severe phenotypic effects due to reduced SAP102 may be due to functional compensation by other MAGUKs. Indeed, PSD-95 depletion does not result in defects in dendritic spine density in primary hippocampal cultures due to compensatory mechanisms by PSD-93. SAP102 has been shown to compensate for some defects in PSD-93/PSD-95 knockout neurons.8, 13, 14
One frame-shift insertion, two nonsense and five splice-site variants have been previously identified in DLG3 in individuals with ID.5, 10, 11, 12 Most of these variants introduce a premature stop codon within or before the PDZ3 domain and potentially lead to a truncation or absence of SAP102 protein5, 10, 11, 12 (Supplementary Figure 5). DLG3 is among the top 0.3% dosage-sensitive genes in the genome, suggesting these variants are most likely pathogenic.15 Affected individuals presented either mild to moderate ID with delayed speech development10 or moderate to severe ID with behavioural problems.12 Compared with the known loss-of-function variants, the DLG3 5ʹ UTR G duplication that we identified would result in reduced, but not total loss of SAP102 protein, which is consistent with the reduced severity of phenotype in our family compared with previous reports.
Our dual luciferase reporter assays and patient versus control LCL DLG3 mRNA and protein expression data showed that the DLG3 5ʹ UTR G duplication results in reduced levels of SAP102 protein but not mRNA. Such observations can be tested using neurons derived from induced pluripotent stem cells (iPSCs) of the affected individuals and controls, however, these iPSCs were not available. We argue that reduced SAP102 in the affected LCLs and luciferase reporter activity from the G duplication in the DLG3 5ʹ UTR is post transcriptional and most likely due to one or more mRNA structural alterations that interfere with efficient translation initiation. We observed that the G duplication was predicted to result in significant alteration to the DLG3 5ʹ UTR mRNA folding, an effect not seen for other ExAC variants, deletion of G in the same set of Gs, and interestingly, also in shorter DLG3 5ʹ UTR sequences. Translation is regulated via multiple 5ʹ and 3ʹ UTR structural features such as 5ʹ-cap, secondary structures, internal ribosome entry sites (IRESs), AUG flanking sequences, polyadenylation signals and G-quadruplexes.16, 17 Variants in these elements, particularly those effecting 5ʹ and 3ʹ UTR mRNA secondary structures have been linked to disease.17, 18, 19, 20
Taken together, our combined genetic, experimental and modelling evidence suggests that NC_000023.10:g.69665044dupG variant alters SAP102 protein abundance, which is the most plausible cause of moderate non-syndromic ID this family.
References
Gecz J, Shoubridge C, Corbett M : The genetic landscape of intellectual disability arising from chromosome X. Trends Genet 2009; 25: 308–316.
Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347.
Hu H, Haas SA, Chelly J et al: X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 2015; 21: 133–148.
Huang L, Jolly LA, Willis-Owen S et al: A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am J Hum Genet 2012; 91: 694–702.
Tarpey P, Parnau J, Blow M et al: Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet 2004; 75: 318–324.
Zhang L, Jie C, Obie C et al: X chromosome cDNA microarray screening identifies a functional PLP2 promoter polymorphism enriched in patients with X-linked mental retardation. Genome Res 2007; 17: 641–648.
Grant SG : Synaptopathies: diseases of the synaptome. Curr Opin Neurobiol 2012; 22: 522–529.
Cuthbert PC, Stanford LE, Coba MP et al: Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci 2007; 27: 2673–2682.
Murata Y, Constantine-Paton M : Postsynaptic density scaffold SAP102 regulates cortical synapse development through EphB and PAK signaling pathway. J Neurosci 2013; 33: 5040–5052.
Philips AK, Siren A, Avela K et al: X-exome sequencing in Finnish families with intellectual disability--four novel mutations and two novel syndromic phenotypes. Orphanet J Rare Dis 2014; 9: 49.
Tzschach A, Grasshoff U, Beck-Woedl S et al: Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet 2015; 23: 1513–1518.
Zanni G, van Esch H, Bensalem A et al: A novel mutation in the DLG3 gene encoding the synapse-associated protein 102 (SAP102) causes non-syndromic mental retardation. Neurogenetics 2010; 11: 251–255.
Oliva C, Escobedo P, Astorga C, Molina C, Sierralta J : Role of the MAGUK protein family in synapse formation and function. Dev Neurobiol 2012; 72: 57–72.
Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA : Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron 2006; 52: 307–320.
Huang N, Lee I, Marcotte EM, Hurles ME : Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 2010; 6: e1001154.
Bugaut A, Balasubramanian S : 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res 2012; 40: 4727–4741.
Chatterjee S, Pal JK : Role of 5'- and 3'-untranslated regions of mRNAs in human diseases. Biol Cell 2009; 101: 251–262.
Damjanovich K, Langa C, Blanco FJ et al: 5'UTR mutations of ENG cause hereditary hemorrhagic telangiectasia. Orphanet J Rare Dis 2011; 6: 85.
Nicolas G, Wallon D, Goupil C et al: Mutation in the 3' untranslated region of APP as a genetic determinant of cerebral amyloid angiopathy. Eur J Hum Genet 2015; 24: 92–98.
Simon D, Laloo B, Barillot M et al: A mutation in the 3'-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum Mol Genet 2010; 19: 2015–2027.
Acknowledgements
Participation of the affected family in the study and funding support from National Health and Medical Research Council (APP1008077 and APP44107859 grants) and WCH foundation grants (MC, DP, RK and JG) and MS McLeod Research Fellowship (MC) are thankfully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
In memoriam to Dr Michael Partington, who worked with this family for many years and unfortunately did not get to see this outcome.
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kumar, R., Ha, T., Pham, D. et al. A non-coding variant in the 5ʹ UTR of DLG3 attenuates protein translation to cause non-syndromic intellectual disability. Eur J Hum Genet 24, 1612–1616 (2016). https://doi.org/10.1038/ejhg.2016.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.46
This article is cited by
-
Identification of a DLG3 stop mutation in the MRX20 family
European Journal of Human Genetics (2024)
-
X-chromosomale Intelligenzminderung
Medizinische Genetik (2018)