Abstract
Alexander disease (AxD) is a fatal neurodegenerative disorder of astrocyte dysfunction in man, for which already a number of causal variants are described, mostly de novo dominant missense variants in the glial fibrillary acidic protein (GFAP). A similar disorder was already phenotypically described in animals but without the identification of causal variants. We diagnosed a Labrador retriever with a juvenile form of AxD based on clinical (tetraparesis with spastic front limbs mimicking ‘swimming puppy syndrome’) and pathological (the detection of GFAP containing Rosenthal fibers in astrocytes) features. In order to identify a causal variant, the coding sequences of the four detected GFAP transcript variants (orthologues from human transcript variants α, γ, δ/ɛ and κ) were sequenced. From the five detected variants, a heterozygous c.719G>A nucleotide substitution resulting in a p.Arg240His substitution was considered to be causal, because it is orthologous to the heterozygous de novo dominant c.716G>A (p.Arg239His) hotspot variant in man, proven to cause a severe phenotype. In addition, the variant was not found in 50 unrelated healthy Labrador retrievers. Because the condition in dogs is morphologically similar to man, it could be a promising animal model for further elucidating the genotype/phenotype correlation in order to treat or prevent this disease.
Similar content being viewed by others
Introduction
Astrocytes, the most numerous cell type of the central nervous system, use their dynamic intermediate filament network, which mainly consists of glial fibrillary acidic protein (GFAP; Gene/Locus MIM number 137780), as a signalling platform and a structural scaffold to coordinate appropriate responses in health and disease.1 Similar to other intermediate filament proteins, GFAP consists of an N-terminal head (random coil), a highly conserved central rod domain (subdivided into four α-helices and separated by three short non-helical linkers) and a C-terminal tail (random coil).2 The human GFAP gene is located on chromosome 17q21 and its main isoform α comprises 9 exons and 432 amino acids. The other nine described GFAP isoforms differ mainly in rod domain length and tail sequence and are differentially expressed by specific astrocyte subpopulations.1 Overexpression, isoform ratios and variants in the rod domain and the tail of GFAP all influence the intermediate filament network structure and function.3
Alexander disease4 (AxD; Phenotype MIM number 203450) is a progressive, lethal neurodegenerative astrocyte dysfunction in man and was originally classified based on the age of onset as infantile, juvenile and adult (reviewed in Brenner et al5 and Messing et al6). Recently, Prust et al7 proposed a reclassification in two age-dependent clinical subtypes: type I, characterized by an early age of onset, seizures, macrocephaly, encephalopathy, developmental delay, paroxysmal deterioration, failure to thrive and typical magnetic resonance imaging (MRI) features, and type II, characterized by a later age of onset, autonomic dysfunction, bulbar symptoms, ocular movement abnormalities and atypical MRI features. The characteristic pathological feature of both types of AxD are widespread and abundant astrocytic protein aggregates, termed Rosenthal fibers (RFs),8 that mainly contain GFAP.
Until now, all known genetic causes of AxD are attributed to GFAP variants (explaining >95% of the cases), mostly de novo dominant missense variants creating amino-acid substitutions at the p.Arg79 and p.Arg239 hotspots.7, 9 Because most identified variants generate practically full-length GFAP, GFAP null mice do not display signs of AxD and transgenic mice overexpressing GFAP develop a fatal encephalopathy with accumulations of GFAP in RFs, it is believed that variants cause disease by a dominant gain of function.6
A similar disorder (OMIA 001208-9615) was already phenotypically described in 12 dogs (5 Bernese mountain dogs, 2 Labrador retrievers, 1 Bernese mountain crossbred dog, 1 Chihuahua, 1 French bulldog, 1 Miniature poodle and 1 Scottish terrier dog)10, 11, 12, 13, 14, 15, 16, 17 and 4 sheep (3 Merino sheep and 1 White alpine sheep),18, 19 but until now no causal variants were described.
Here we describe the clinical and pathological features of another Labrador retriever diagnosed with AxD and the identification of the causal GFAP variant by sequencing the coding regions of the expressed transcript variants.
Materials and methods
Clinical examination
A 3-month-old Labrador retriever was presented to our clinic for the investigation of progressively worsening tetraparesis. Clinical and neurological examinations were performed, together with a complete blood work, radiography of the thorax, electrophysiological examination and a cisternal cerebrospinal fluid tap. Additionally, the dog was tested for hereditary centronuclear myopathy (OMIA 001374-9615) and Neospora in the blood.
Pathological examination
A necropsy was performed. Samples from the brain were immediately frozen. Additional samples from the brain, spinal cord and other organs were formalin fixed and routinely processed for histopathological evaluation. Five-μm sections of the samples were stained with haematoxylin and eosin. Immunohistochemical staining for GFAP was performed on the samples of the central nervous system (Dako, Cat. no. K4011, Glostrup, Denmark). Therefore, sections were dewaxed in xylene, rehydrated through alcohols and subjected to antigen retrieval using proteinase K. Any endogenous peroxidase activity was quenched by incubating with hydrogen peroxide. The slides were incubated with a polyclonal rabbit anti-GFAP antibody (Dako, Cat. no. Z0334), dilution 1:2000. Next, a horseradish peroxidase-labelled polymer, which is conjugated with secondary antibodies, was applied. The immunoreactions were visualized using 3,3’-diaminobenzidine as chromogen and counterstained with haematoxylin. Tissue sections from the brain of a dog that died from an unrelated cause were included as control material.
Genetic analysis
Genomic DNA was isolated from 100 mg frozen brain tissue using proteinase K digestion, phenol/chloroform extraction and ethanol precipitation.20 DNA quality and quantity was assessed with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). PCR was performed with FastStart DNA Polymerase (Roche Diagnostics, Mannheim, Germany) on 20 ng pure (OD260/280=2.0) genomic DNA.
Total RNA was isolated from 100 mg frozen brain tissue using the Aurum Total RNA Fatty and Fibrous Tissue Kit, including an on-column DNase digest (Bio-Rad Laboratories, Hercules, CA, USA). RNA quality and quantity was assessed with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and RNA integrity was assessed by evaluating the 28S/18S rRNA bands on gel and possible DNA contamination with minus-RT-PCR. Reverse transcription was performed on 1 μg pure, non-degraded, DNA-free total RNA using the ImProm-II Reverse Transcription System (Promega Corporation, Madison, WI, USA) with oligo(dT)15 and random primers. Complementary DNA was 10 times diluted with water, and 2 μl was used as a template for PCR.
Primer pairs (PP) were designed with Primer3Plus21 based on canine GFAP reference sequence (Acc. no. NC_006591.3) taking into account described sequence variants (dbSNP22), repeat sequences (RepeatMasker23) and secondary structures (Mfold24). Specificity of the primers was checked via ePCR using BiSearch.25 PCR amplicons were analysed via agarose gel electrophoresis and eluted via the GeneClean II Kit (MP Biomedicals, Santa Ana, CA, USA). Sequencing reactions were performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), with the individual PCR primers as sequencing primers and analysed at Eurofins Genomics (Ebersberg, Germany). Sequence analysis was performed with Unipro UGENE v1.16.1.26 Details on primers/amplicons are given in Tables 1 and 2, and on PCR/sequencing mixes/programs in Supplementary Information File S1.
Results
Clinical features
Since the age of 3 months, the dog developed progressively worsening tetraparesis with a spastic swimming-puppy-like position of the thoracic limbs (see video, Supplementary Information File S2) and a flattened chest. The owner mentioned also some regurgitation. One month later, mild vestibular signs and myoclonic jerks at the head and cervical region became obvious. General clinical examination was within normal limits. Neurological examination revealed absent patellar reflexes, weakness on the four limbs with an abnormal spasticity of the thoracic limbs and mild generalized muscle atrophy. During the second visit, a vestibular strabismus in the right eye, a mild right-sided head tilt and regular myoclonic jerks in the region of the head, neck and thoracic limbs were noticed. The tetraparesis progressively evolved into a non-ambulatory state.
Blood examination, electrophysiological examination and cerebrospinal fluid analysis were within normal limits. Radiography of the thorax confirmed the dorsoventral flattening of the thorax. Because of the worsening of neurological signs, further examinations (eg, MRI of the brain) were declined by the owner, and the pup was euthanized at the age of 4.5 months.
The litter consisted of seven pups of which one was stillborn. One other pup appeared to be clinically normal and both parents are still alive at the time of writing of this manuscript (15 July 2015). Further information about the other littermates was not available.
Pathological features
Except for the dorsoventral flattening of the thorax, no gross abnormalities were noted at necropsy. On histopathological examination, lesions were confined to the central nervous system. In the white and grey matter of the brain, blood vessels were surrounded by numerous short, perpendicular orientated, hypereosinophilic, amorphous intra-astrocytic RFs (Supplementary Information File S3A). These RFs were also found in the astrocytic endfeet in the subpial tissue and in some subventricular locations. All RFs were strongly immunopositive for GFAP (Supplementary Information File S3B). There was proliferation of abnormal astrocytes with large nuclei, prominent nucleoli and glassy eosinophilic to pale cytoplasm. All neuronal populations as well as oligodendroglial nuclei showed no abnormalities.
At all levels of the spinal cord, RFs were found in both white and grey matter, mostly perivascular and subpial. Moderate numbers of hypertrophied astrocytes were noted.
GFAP transcript variant expression
Because most AxD-causing variants in man are GFAP missense variants and the annotation of the GFAP gene and its transcript variants in dog was only predicted in silico, we first investigated experimentally which transcript variants were expressed in the brain of the case. Transcript variant-specific RT-PCR assays were designed based on the 10 described transcript variants in man/mouse/rat and the 3 in silico predicted transcript variants in dog (see Tables 1 and 2, and Supplementary Information File S1 for details).
PP2 amplified only a fragment of 789 bp, indicating that transcript variants α, β and/or γ were expressed and that transcript variants ζ, Δ135, Δ164, Δex6, Δex7 and X4 were not expressed. Transcript variant α (canonical form) and β have the same coding sequence. Because of the lack of knowledge about their different 5′-UTR region, we could not differentiate between the expression of both forms. PP3 confirmed the expression of transcript variant α/β and PP4 the expression of transcript variant γ. PP5 was used to confirm that at least 185 bp of the 3′-side of intron 1 was included in transcript variant γ. PP6 amplified both specific fragments for transcript variants δ/ɛ and κ, confirming their expression (the identity of all fragments was verified by sequencing).
GFAP variant detection
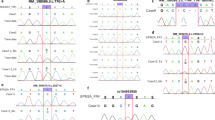
In addition to PP2-6, which amplify all coding sequences represented in the four expressed GFAP transcript variants (α, γ, δ/ɛ and κ; Figure 1) except for exon 1, we amplified and sequenced exon 1 with PP7 and compared all sequences with the predicted canine sequences. All four canine GFAP transcript variant sequences described here were submitted to GenBank (α: Acc. no. KR827462, γ: Acc. no. KT192567, δ/ɛ: Acc. no. KR827463 and κ: Acc. no. KR827464).
Schematic representation of the canine GFAP structure, the exons of the four expressed transcript variants in the brain of the case and the five discovered variants. The canine GFAP structure contains a N-terminal head (random coil), a highly conserved central rod domain subdivided into four α-helices (1A, 1B, 2A and 2B) and separated by three short non-helical linkers (L1, L12 and L2) and a C-terminal tail (random coil). The exons (from start to stop codon) of the four expressed transcript variants α (Acc. no. KR827462), γ (Acc. no. KT192567), δ/ɛ (Acc. no. KR827463) and κ (Acc. no. KR827464) are represented as white blocks with the intervening introns as black lines. The five variants are represented as grey lines (Acc. no. ss1791785830:181G>A, ss1791785831:357C>G, ss1791785832:719G>A, ss1791785833:1224C>T and ss1791785834:1244C>T).
The five detected sequence variants (Figure 1) were submitted to the dbSNP (Acc. no. ss1791785830:181G>A, ss1791785831:357C>G, ss1791785832:719G>A, ss1791785833:1224C>T and ss1791785834:1244C>T). The nucleotide numbering is relative to the start codon in the submitted transcript variant sequences mentioned above.
From the five detected variants, the heterozygous variant c.719G>A in exon 4 encoding p.Arg240His in α-helix 2A was found to be causal, because the exact variant was also proven to be a causal hotspot variant of one of the most severe forms of AxD in 11 human patients (ClinVar:NM_002055.4(GFAP):c.716G>A (p.Arg239His)).7, 9 This finding was added as a comment in the ClinVar entry and was supported by the fact that the causal variant was not found in 50 unrelated healthy Labrador retrievers (assay performed with PP8).
Discussion
The clinical signs of this dog reflected the diffuse involvement of the central nervous system. There was an obvious neurological dysfunction of the motor system that involved all limbs, although the thoracic limbs were clinically more affected than the hind limbs. Attempts to ambulate resulted in a swimming-like motion owing to severe abduction and spasticity of the thoracic limbs. A spasticity of the thoracic limbs and a similar swimming way of motion with the four limbs has been described previously in dogs with AxD.11, 16 The myoclonic jerks, which were noticed with increasing frequency mainly in the facial, cervical and thoracic limb region of our dog, probably were focal seizures, although electroencephalography was not performed. The mild vestibular signs and regurgitation also reflected involvement of the brainstem.
In man, AxD has been classified into three types (infantile, juvenile and adult form) based on the age of onset and the clinical signs,27 although another classification system (type I and type II AxD), based on clinical signs and MRI findings, has been recently proposed.7 The age of onset (3 months) and clinical signs of our dog (eg, spasticity, regurgitation, mild vestibular signs) seem most comparable with the juvenile type of AxD in man. Also, the lack of mental and sensory dysfunction and the absence of megalencephaly points more towards a juvenile type of AxD,27 although it is not easy to reliably assess the mental and developmental state of such a young dog. In this dog, MRI of the brain was declined by the owner, and therefore, it is difficult to apply the more recent classification system.7 However, the early onset and the severity of the neurological signs point towards a type I AxD. Lesions found in the brain and spinal cord of this dog were typical of AxD. Moreover, immunohistochemistry confirmed the accumulation of GFAP in the RFs.
Transcript variant-specific RT-PCR was used as a quick and easy tool to identify the transcribed GFAP exons that had to be targeted for variant detection. Although new experimental evidence was gathered about the identity and the content of the expressed transcript variants, no conclusions could be made about the relative transcript variant expression levels based on this end point detection method.
Transcript variant α was correctly predicted in dog as transcript variant X2. It shows 89% sequence identity on DNA level and 94% on the protein level with its human orthologue. The canine orthologue is 1 amino-acid longer due to a p.Tyr14_Val15dup and a p.Leu46del, both located in the head region and bordering the least conserved region of the canonical protein. As a result, the orthologous causal variant is termed c.716G>A (p.Arg239His) in man and c.719G>A (p.Arg240His) in dogs.
The 5′-side of transcript variant γ (not expressed in reactive astrocytes in man) is still unknown.1 It lacks exon 1 and only the last 126 bp of intron 1 (not containing an in-frame start codon) are known to be part of the human transcript. In dogs, we showed that at least the last 185 bp of intron 1 are included in the transcript, containing an in-frame start codon followed by three in-frame stop codons, raising the question what could be the (regulatory?) function of this transcript.
Transcript variant δ/ɛ, not predicted in dogs, shows 89% sequence identity on DNA level and 92% on protein level with its human orthologue.
Transcript variant κ, correctly predicted in dogs as transcript variant 3, shows 88% sequence identity on DNA level and 92% on protein level with its human orthologue until the c.1235delG in exon 7b. After that position (encoded in the 3′-part of the tail), there is no conservation with any described orthologue.
All other transcript variants were not detected, including the canine in silico predicted transcript variant X4, which is neither found in man, mouse nor rat but is predicted in pig, horse and cow.
Variants c.181G>A, c.357C>G and c.1224C>T were not considered to be causal, because the first one was also found in the reference sequence of pig, horse and cat and the latter two were silent variants. Variant c.1244C>T (p.Ser415Phe) was found in exon 7b of transcript variant κ. This missense variant could potentially influence the function of this isoform, but because it is located in the non-conserved tail region, it is impossible to draw any conclusion at this point.
We concluded that the heterozygous GFAP variant c.719G>A (p.Arg240His) is the causal variant, because the exact variant also exists in man as the heterozygous de novo dominant c.716G<A (p.Arg239His) hotspot variant and was already proven to cause one of the most aggressive AxD phenotypes in 11 unrelated patients.7, 9 In addition, the variant was not found in 50 unrelated healthy Labrador retrievers. Because the owners of the parents of the case did not want to participate in this study, we could not confirm if the variant was present or not in the parents or littermates. However, because we know that both parent dogs are still alive and healthy, and taking into account the severe phenotype present in this pup as in the human patients carrying the orthologous variant, we conclude that this variant is a de novo variant.
In man with AxD, genotype–phenotype correlations still have to be determined and a clear distinction between the different types of AxD is not always clear.28 Because the condition in dogs is morphologically and genetically similar to man, it could be a promising animal model for further elucidating the genotype/phenotype correlation in order to treat or prevent (application of drugs or gene therapy) this disease in man. Although the disorder is quite rare, understanding how astrocyte dysfunction contributes to the phenotype in AxD and how astrocyte function might be restored will also contribute to the studies of more common neurological diseases (such as epilepsy, Alzheimer's, Parkinson's and Huntington's disease) or ageing.
In addition, this animal model could also be used to further investigate the mechanism by which the same de novo variants are created in unrelated individuals of different species. Although variants are still often considered as a result of a random process, it is shown that certain regions are inherently variant prone by virtue of their local DNA sequence environment or by higher order features of the genomic architecture.29 The causal variant described here probably arose because it is part of a CpG dinucleotide variant hotspot.5 Unravelling these mechanisms might open the way to predict the creation of disease-causing de novo variants.
References
Hol EM, Pekny M : Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr Opin Cell Biol 2015; 32: 121–130.
Sawaishi Y : Review of Alexander disease: beyond the classical concept of leukodystrophy. Brain Dev 2009; 31: 493–498.
Kamphuis W, Middeldorp J, Kooijman L et al: Glial fibrillary acidic protein isoform expression in plaque related astrogliosis in Alzheimer's disease. Neurobiol Aging 2014; 35: 492–510.
Alexander WS : Progressive fibrinoid degeneration of fibrillary astrocytes associated with mental retardation in a hydrocephalic infant. Brain 1949; 72: 373–381.
Brenner M, Goldman JE, Quinlan RA, Messing A : Alexander disease: a genetic disorder of astrocytes; in Parpura V, Haydon PG (eds): Astrocytes in (Patho)Physiology of the Nervous System. Springer: New York, NY, USA, 2009, pp 591–648.
Messing A, Brenner M, Feany MB, Nedergaard M, Goldman JE : Alexander disease. J Neurosci 2012; 32: 5017–5023.
Prust M, Wang J, Morizono H et al: GFAP mutations, age at onset, and clinical subtypes in Alexander disease. Neurology 2011; 77: 1287–1294.
Rosenthal W : Uber eine eigenthumliche, mit syringomyelie complicirte geschwulst des ruckenmarks. Bietr Pathol Anat 1898; 23: 111–143.
Alexander Disease. Website Waisman Center, University of Wisconsin-Madison. https://www.waisman.wisc.edu/alexander-disease/ (last updated 13 July 2015).
McGrath JT : Fibrinoid leukodystrophy (Alexander’s disease); in Andrews EJ, Ward BC, Altman NH (eds): Spontaneous Animal Models of Human Disease. Academic Press: New York, NY, USA, 1979, Vol 2, pp 147–148.
Cox NR, Kwapien RP, Sorjonen DC, Braund KG : Myeloencephalopathy resembling Alexander’s disease in a Scottish terrier dog. Acta Neuropathol (Berl) 1986; 71: 163–166.
Richardson JA, Tang K, Burns DK : Myeloencephalopathy with Rosenthal fiber formation in a Miniature Poodle. Vet Pathol 1991; 28: 536–538.
Weissenböck H, Obermaier G, Dahme E : Alexander’s disease in a Bernese Mountain dog. Acta Neuropathol (Berl) 1996; 91: 200–204.
Aleman N, Marcaccini A, Espino L, Bermudez R, Nieto JM, Lopez-Pena M : Rosenthal fiber encephalopathy in a dog resembling Alexander disease in humans. Vet Pathol 2006; 43: 1025–1028.
Ito T, Uchida K, Nakamura M, Nakashima K, Suzuki K, Nakayama H : Fibrinoid leukodystrophy (Alexander's disease-like disorder) in a young adult French bulldog. J Vet Med Sci 2010; 72: 1387–1390.
Gruber A, Pakozdy A, Leschnik M, Mai S, Weissenböck H : Morbus Alexander - 4 Fälle bei Hunden in Österreich. Wien Tierärztl Mschr 2010; 97: 298–301.
Wrzosek M, Giza E, Płonek M, Podgórski P, Vandevelde M : Alexander disease in a dog: case presentation of electrodiagnostic, magnetic resonance imaging and histopathologic findings with review of literature. BMC Vet Res 2015; 11: 115.
Fankhauser R, Fatzer R, Bestetti G, Deruaz JP, Perentes E : Encephalopathy with Rosenthal fiber formation in a sheep. Acta Neuropathol (Berl) 1980; 50: 57–60.
Kessell AE, Finnie JW, Manavis J, Cheetham GD, Blumbergs PC : A Rosenthal fiber encephalomyelopathy resembling Alexander's disease in 3 sheep. Vet Pathol 2012; 49: 248–254.
Sambrook J, Fritsch EF, Maniatis T eds: Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM : Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35: W71–W74.
Sherry ST, Ward MH, Kholodov M et al: dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311.
Smit AFA, Hubley R, Green P RepeatMasker Open-4.0. 2013-2015 (<http://www.repeatmasker.org>).
Zuker M : Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31: 3406–3415.
Arányi T, Váradi A, Simon I, Tusnády GE : The BiSearch web server. BMC Bioinformatics 2006; 7: 431.
Okonechnikov K, Golosova O, Fursov M : the UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 2012; 28: 1166–1167.
Springer S, Erlewein R, Naegele T et al: Alexander disease – classification revisited and isolation of a neonatal form. Neuropediatrics 2000; 31: 86–92.
Asahina N, Okamoto T, Sudo A, Kanazawa N, Tsujino S, Saitoh S : An infantile-juvenile form of Alexander disease caused by a R79H mutation in GFAP. Brain Dev 2006; 28: 131–133.
Cooper DN, Bacolla A, Férec C, Vasquez KM, Kehrer-Sawatzki H, Chen JM : On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Hum Mutat 2011; 32: 1075–1099.
Feinstein DL, Weinmaster GA, Milner RJ : Isolation of cDNA clones encoding rat glial fibrillary acidic protein: expression in astrocytes and in Schwann cells. J Neurosci Res 1992; 32: 1–14.
Zelenika D, Grima B, Brenner M, Pessac B : A novel glial fibrillary acidic protein mRNA lacking exon 1. Brain Res Mol Brain Res 1995; 30: 251–258.
Acknowledgements
We thank Julie Van de Putte, Dominique Vander Donckt, Linda Impe, Ruben Van Gansbeke and Delphine Ameye for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Van Poucke, M., Martlé, V., Van Brantegem, L. et al. A canine orthologue of the human GFAP c.716G>A (p.Arg239His) variant causes Alexander disease in a Labrador retriever. Eur J Hum Genet 24, 852–856 (2016). https://doi.org/10.1038/ejhg.2015.223
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.223
This article is cited by
-
Symptomatic care of late-onset Alexander disease presenting with area postrema-like syndrome with prednisolone; a case report
BMC Pediatrics (2022)
-
Truncating SLC12A6 variants cause different clinical phenotypes in humans and dogs
European Journal of Human Genetics (2019)