Abstract
In recent years, several association analysis methods for case-control studies have been developed. However, as we turn towards the identification of single nucleotide polymorphisms (SNPs) for prognosis, there is a need to develop methods for the identification of SNPs in high dimensional data with survival outcomes. Traditional methods for the identification of SNPs have some drawbacks. First, the majority of the approaches for case-control studies are based on single SNPs. Second, SNPs that are identified without incorporating biological knowledge are more difficult to interpret. Random forests has been found to perform well in gene expression analysis with survival outcomes. In this paper we present the first pathway-based method to correlate SNP with survival outcomes using a machine learning algorithm. We illustrate the application of pathway-based analysis of SNPs predictive of survival with a data set of 192 multiple myeloma patients genotyped for 500 000 SNPs. We also present simulation studies that show that the random forests technique with log-rank score split criterion outperforms several other machine learning algorithms. Thus, pathway-based survival analysis using machine learning tools represents a promising approach for the identification of biologically meaningful SNPs associated with disease.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have enormous potential in identifying new susceptibility genes for complex disease. However, the high-dimensional nature of GWAS data makes it challenging to distinguish true signals from background noise. Most published studies have looked at single locus comparisons to identify single nucleotide polymorphisms (SNPs) between cases and controls. By taking the one SNP at a time approach, GWAS studies may be underpowered to detect smaller effects. There is increasing evidence to suggest that gene–gene interactions may have a role in the etiology of complex disease. By analogy with high-dimensional microarray data analysis, several methods have been proposed for incorporating previous information in genome-wide association analysis. Chasman et al, Peng et al, and Ritchie et al have advocated the use of previous knowledge for GWAS analyses and have described the advantages of using pathway-based methods.1, 2, 3 Baranzini et al have implemented this approach in the analysis of multiple sclerosis by using a network-based analysis to tease out SNPs with association P-values between 0.05 and 10−8 in the original single-SNP association analysis.4 Ballard et al performed two pathway-based tests, a binomial test and a random set method to identify pathways associated with rheumatoid arthritis.5 Wang et al utilized a modified gene set enrichment method for SNP data to identify pathways associated with Crohn's disease.6 Both Wang et al and Dinu et al performed pathway-based analyses to study age-related macular degeneration.7, 8 The former paper also investigated two GWAS of Parkinson's disease. However, little has been done to apply pathway-based methods to correlate SNP data with survival outcomes.
Random forests classification has been applied to identify SNPs associated with binary outcomes in a pathway-based setting.9, 10, 11 Overall, random forests is among the best approaches for analyzing survival time using gene expression data.12, 13, 14 In this article, we introduce one of the first methods to correlate SNP with survival outcomes. We compare two different implementations of random forests for survival outcomes and other machine learning approaches through simulations. Moreover, we illustrate the use of our pathway-based method for survival SNP analysis through application to a multiple myeloma data set and investigate how linkage disequilibrium (LD) may affect prediction. In summary, random survival forests with log-rank score (LRS) split performed best in both simulation and real data analysis. We were able to identify two pathways that are associated with survival outcomes of interest in multiple myeloma patients.
Materials and methods
Several machine learning methods are compared in identifying pathways associated with SNP data. We describe below random survival forests, which performed among the best in simulations.15 Several other machine learning methods are presented in Supplementary materials. The goal of these machine learning methods is to identify pathways containing SNPs that can predict the survival outcome of the population of interest.
Random survival forests
For SNP data, we code each individual SNP as values 0, 1, and 2 for the number of variant alleles at the respective SNP. The random forests method for survival outcome was first proposed by Leo Breiman (http://oz.berkeley.edu/users/breiman/SF_Manual.pdf). It has since been refined with different variations. One of the popular variants is random survival forests.15 A random survival forests encompasses many binary trees, each of which is formed by a deterministic algorithm. First, a best binary split is chosen using a subset of SNPs within a pathway. Second, every tree is built using a bootstrap sample of the patients of interest. Unlike classification and regression trees (CART), no pruning is involved. Several split criteria are available in random survival forests; we apply the log-rank and LRS for split criteria as described below. Other split criteria such as conserve and random are given in Supplementary materials.
The RSF algorithm is applied as follows. First, bootstrap samples are drawn from the original data ntree times, where ntree is the number of trees. For each bootstrap sample, some samples are left out-of-bag (OOB). A binary survival tree is grown for each bootstrap sample. Let p be the number of SNPs in a pathway. At each node of the tree, p1/2 SNPs in the pathway are selected at random for splitting. Using one of the split criteria described below, a node is split using a single SNP from the p1/2 randomly chosen SNPs that maximizes the survival differences between the children nodes. The splitting continues until each terminal node reaches the minimum number of events with unique survival times. The default is three for right censored data.15 Next, binary survival trees are aggregated to obtain the ensemble cumulative hazard estimates, which will also be detailed below.
Let i denote an individual with i=1, … , n, let n be the total number of individuals and let x be one of the SNP predictors. Splits are of form x ≤c and x >c, where c is the cutoff value. Let n1=  , an indicator function counting the number of observations less than or equal to the cutoff. Split criterion LRS,, which measures the node separation, is based on the log-rank test statistic, and uses the following equation:
, an indicator function counting the number of observations less than or equal to the cutoff. Split criterion LRS,, which measures the node separation, is based on the log-rank test statistic, and uses the following equation:

where  , and Ii=1 if an event is observed for individual i and 0 otherwise, and
, and Ii=1 if an event is observed for individual i and 0 otherwise, and  ; and μa and sa2 are the sample mean and sample variance of ai, respectively.16 The best split is defined as the one that maximizes the absolute value of the equation LRS(X,c) above.
; and μa and sa2 are the sample mean and sample variance of ai, respectively.16 The best split is defined as the one that maximizes the absolute value of the equation LRS(X,c) above.
Another split criterion is the log-rank test (LR) criterion, which measures the node separation, and is defined as:

where E is the number of distinct event times T(1)≤T(2) ≤…≤ T(E) in the parent node;  is the number of events at time ti in the child nodes j=1,2;
is the number of events at time ti in the child nodes j=1,2;  is the number of individuals at risk at time ti in the child nodes j=1,2; and
is the number of individuals at risk at time ti in the child nodes j=1,2; and  and
and  .17 Again, the best split is chosen similarly, that is, it maximizes the absolute value of the equation LR(X,c) above.
.17 Again, the best split is chosen similarly, that is, it maximizes the absolute value of the equation LR(X,c) above.
Trees are aggregated to form the forest through the ensemble cumulative hazard function (eCHF), which groups the hazard estimates from the terminal nodes. The CHF estimates for a terminal node L is the Nelson-Aalen estimator

where ti,L = distinct survival time;  =the number of events; and
=the number of events; and  =the number of individuals at risk at time (ti,L). For every binary survival tree with Q terminal nodes, there will be Q different CHF estimators.
=the number of individuals at risk at time (ti,L). For every binary survival tree with Q terminal nodes, there will be Q different CHF estimators.
The CHF estimate for an individual inew with SNP predictor snpnew can be found by identifying which terminal node includes the individual when it is dropped down the binary survival tree. That is, the CHF estimate is equal to  if inew is found in terminal node L. The ensemble CHF is simply the sum of the CHFs across the bootstrap samples divided by number of trees. The expected number of ensemble events can be obtained by summing over time Tj for j=1 to n. The description of support vector machine approaches for survival outcomes, Cox boosting, conditional inference survival forests, and two other split criteria for random survival forests are given in Supplementary materials.
if inew is found in terminal node L. The ensemble CHF is simply the sum of the CHFs across the bootstrap samples divided by number of trees. The expected number of ensemble events can be obtained by summing over time Tj for j=1 to n. The description of support vector machine approaches for survival outcomes, Cox boosting, conditional inference survival forests, and two other split criteria for random survival forests are given in Supplementary materials.
Identification of pathways associated with survival
Our goal is to test whether specific sets of SNPs from the same pathway are strong prognostic factors. One way to do this is to find the expected survival times and expected number of events from machine learning methods, such as random survival forests. The expected number of events is then split for the two groups into approximately equal sizes of high and low survival times or events. We can then compute a log-rank test to see whether there is a significant difference between the high- and low-risk groups. The expected survival times and number of events are obtained using 10-fold cross-validation. At each of the k-fold iteration, 90% of the training data are used to build the random forests model for survival data. The remaining 10% is then used to make predictions on testing individuals who are not involved in training the model. To clarify, the high-risk group has a higher expected number of events from the 10-fold cross-validation prediction compared with the median among all patients, whereas the low-risk group has fewer than or the same number as the median. A small P-value would indicate that this set of SNPs is informative about the prognosis of patients and pathways can be ranked according to the P-values to assess the relative importance of the pathways.
Selection of important SNPs
In addition to identifying pathways, random survival forests can also pick out SNPs from top pathways that are associated with the survival outcome of interest. There is a built-in feature selection procedure based on variable importance in random survival forests. There are two ways to determine the importance in random survival forests: permutation or random split. They give possible ways to quantify which SNPs are most informative, that is, contribute most to the prediction accuracy, for achieving a sound survival prediction. Finding an informative SNP is an indication of the strength or usefulness of its prognostics capability.
To obtain the importance measure for a SNP in a particular pathway, the random survival forests algorithm permutes the values of the SNP in the OOB cases and the cases with permuted values are dropped down their in-bag survival tree. The CHF is then calculated for each tree and aggregated across the trees. The randomly permuted values of the SNP in the OOB individuals and the outcome of interest are independent of each other. The variable importance for a predictor x is equal to PEo−PEn, where PEo is the prediction error of the original ensemble and PEn is the prediction error of the new ensemble with values of predictor x randomly permuted. If the SNP is a good predictor, the SNP is likely to be close to the origin of the tree and a large proportion of trees will contain the SNP. This implies that we expect a decrease in prediction accuracy compared with the value before the random permutation.
Mapping of SNPs to genes on the pathway
The gene sets are obtained from the Broad Institute (http://www.broad.mit.edu/) and included 203 KEGG,18 and 278 BioCarta pathways (http://www.biocarta.com). SNPs with minor allele frequencies ≥0.05 that are within 5 kb up and downstream from any gene are considered. This results in 154 979 out of 500K SNPs being mapped to pathways. If a SNP is located within shared regions of two overlapping genes, the SNP will be mapped to both genes.
Simulation studies
We next used simulations to evaluate the performance of different methods in the identification of SNPs. For the alternative case, a pathway from the real data set with small P-values was chosen. The genotype data was generated using the multinomial distribution. The probability for the classes, 0, 1 and 2 were taken from the real data to retain the pathway correlation structure. The survival times S were generated as exponentially distributed random variables with the addition of an ɛ distributed as N[0,0.5].19 Under the alternative case, β equals one for the top five informative SNPs and 0 otherwise. The censoring time (CT) was generated as an N(max(S),3), which resulted in censoring of 20–45% of events for each simulated data set. If the generated CT was less than the generated survival time, the survival time for that individual was considered as censored. For the null case, in which β equals 0 for all the predictors, a pathway from the real data set with large P-values was chosen. Each simulation generated 50 multinomial distributed SNPs with sample size 96, 192 or 288.
Imputation
To infer the missing SNP genotypes in the real data set, we impute the non-genotyped markers in our data set by using the HapMap CEU panel release 27 (NCBI build 36) (http://hapmap.ncbi.nlm.nih.gov/) reference panel,20 and BEAGLE software (http://faculty.washington.edu/browning/beagle/beagle.html).21 BEAGLE uses a localized haplotype-cluster model and a hidden Markov model. Intermarker LD is incorporated to create the most parsimonious model. BEAGLE has been found to perform well compared with other publicly available packages.22
Results
To assess the type I error rate, we simulated 1000 data sets from the null hypothesis as described in the previous section. For every simulated data set, we first calculated the LR test P-value from 10-fold cross-validation as described in the Materials and methods section. For both type I error and power, we calculated the ratio between the number of pathways having a P<0.05 and the number of simulated data sets.
Table 1 shows that the observed random forests type I errors were around the nominal 0.05 level across different sample sizes for RSF with LRS split rule (RSF LRS) and SVMsurv. For sample size 288, Cox boosting, RSF with LR split rule (RSF LR) and random, all had slightly inflated type I error. The type I error for cforest was inflated for all sample sizes. Among the random forests methods, random survival forests with LRS split had the smallest or 2nd smallest type I error on the different sample sizes. In terms of power (Table 2), all methods failed to achieve sufficient power when sample size was less than about 100. SVMsurv had the lowest power across all sample sizes. RSF LR, LRS, cforest, Cox boosting do better than other methods, achieving close to 90% power or above with 192 samples and close to 100% for 288 samples. It is not surprising that the Cox boosting algorithm had superior power, as the Cox model was used to create the simulated data set. With consideration of both type I error and power, RSF LRS is the best method given a sample size of around 200 or above under similar correlation structure to the simulated data set.
Applications to multiple myeloma data set
We next applied our method to the GWAS data from Avet-Loiseau et al.23 They performed a genome-wide analysis of malignant plasma cells from 192 multiple myeloma patients,23 using the Affymetrix (Santa Clara, CA, USA) GeneChip Human Mapping 500K Array Set, to identify markers associated with overall survival. They provided insights into how chromosomal aberrations might have prognostic implications for multiple myeloma patients.
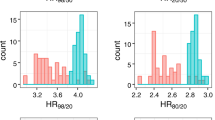
To control for multiple testing, we used the false discovery rate (FDR), with the q-value method.24 Controlling the FDR is one of the preferred methods to adjust for multiple comparisons. The FDR procedure controls the proportion of false positives at a desired level of α, type I errors. Only random survival forests was able to identify pathways that were significant with FDR correction at the 0.1 and 0.05 levels, see Table 3. The FDR cutoff of 0.1 has been commonly used in case-control GWAS studies.25, 26 The pathways significant at this FDR level are cytokine network and stress induction of HSP regulation, see Figures 1, 2. The high- and low-risk groups were determined as defined in the ‘Identification of pathways associated with survival’ section. They contain 79 and 92 SNPs, respectively. For a table with the number of pathways identified based on unadjusted P-values, see Supplementary materials.
Stress induction of HSP regulation is tied to several pathways, including the FAS signaling, mitochondrial, and NF-κB pathways. The NF-κB pathway was hypothesized to have lower expression among high-risk patients.23 The cytokine network pathway has been thoroughly reviewed for prognostic and therapeutics implications in multiple myeloma, and it is also well known that cytokines have a crucial role in the disease etiology of lymphomas.27, 28
To investigate the biological plausibility of our findings, we looked at the informative SNPs in the two pathways. For stress induction of HSP regulation, BCL2 and CASP3 were found to be the most important top 5% SNPs in identifying low- and high-risk survival groups within the pathway, see Table 4 and Table 8 in Supplementary materials (accounting for LD). The protective associations of two caspase genes, CASP3 and CASP9, have been observed and genetic variation in CASP genes has been suggested to be key to the disease etiology of multiple myeloma.29 Novel drugs have been shown to have direct anticancer effects on human myeloma cells, not only by inducing apoptosis via both caspase-dependent and -independent pathways, but also by promoting caspase activation resulting in drug-induced cytotoxicity in multiple myeloma cell lines.30, 31 In addition, genomic region 4q35.1 has recently been identified as a susceptibility locus for chronic lymphocytic leukemia.32 Regarding the BCL2 variant, encouraging results were revealed in phase 1 and 2 studies performed with BCL2 antisense agents and high dose statin with chemotherapy for pretreated myeloma patients.33 Furthermore, the BCL2 locus at 18q21.33 was shown to be frequently amplified in multiple myeloma.34, 35 Additionally, over a decade ago, it was hypothesized that BCL2 has a protective effect in multiple myeloma cells by acting through the NF-κB activation-signaling pathway.36 This pathway was noted above for its links with stress induction of HSP regulation.
For the cytokines network, IL15, IL18, and IL12A were found to be the most important 5% SNPs in the pathway, see Tables 4 and 9 in Supplementary materials (accounting for LD). Several authors have linked IL15 with disease progression in multiple myeloma patients. Jumei et al suggested that IL15 is the primary survival and growth factor for natural killer cells during natural killer lymphopoiesis for relapsed myeloma patients.37 Another research group demonstrated that IL15 contributes to tumor propagation in multiple myeloma.38 Finally, serum IL15 levels have also been found to be elevated in multiple myeloma patients and may be diagnostic for disease progression in multiple myeloma.39 High levels of IL18 in serum have been associated with poor prognosis in multiple myeloma patients.40 A Japanese research group has shown that IL18 inhibits the growth of multiple myeloma cells in the bone marrow and implicated IL18 as a therapeutic target for multiple myeloma.41 Another Japanese research group has further investigated IL18's role in the bone destruction of multiple myeloma patients.42 Finally, IL12A SNPs are associated with an elevated risk of multiple myeloma in a population based case-control study among CT women.43
LD
Previous research has found that intermarker LD does not reduce the predictive power of random forests in the case-control setting.44, 45 We examined whether this holds true for survival outcomes. First, we investigated whether LD among SNPs within a pathway affects the prediction. Pairwise LD was calculated by r2.46 We performed 10 independent 10-fold cross-validation runs with the restriction that SNPs with r2>0.8 were not allowed in the same run for each of the significant pathways (cytokine network and stress induction of HSP regulation). Once SNPs with high LD were removed, the P-values comparing the high-risk and low-risk groups remained highly significant (Supplementary Tables 6 and 7). For the stress induction of HSP regulation and cytokine network pathways, all the P-values were <0.00001 and 0.000001, respectively.
The above approach is similar to the RF1 approach taken by Meng et al under the case-control setting.47 Our cross-validation and random forests survival prediction approach is robust to the presence of SNPs with high LD, in agreement with previous reports.44, 45 The effect of LD on the ranking of the variable importance measure in random forests is presented in Supplementary materials.
Discussion
We have described a pathway-based approach for analyzing SNP data with survival outcome using random survival forests. This approach allows us to identify pathways that are strong predictors of patient's survival. The ability of the SNPs within a pathway in distinguishing high and low-risk groups are tested using a log-rank test. The log-rank test P-values are further adjusted for multiple comparisons using FDR. This approach can help biomedical researchers tease out more biologically meaningful prognostic SNPs from complex GWAS data. We illustrated the use of our approach in a multiple myeloma data set genotyped with the Affymetrix 500K SNP array. We compared random survival forests with other machine learning algorithms including Cox boosting, support vector machine for survival and conditional inference survival forest. Random survival forest with LRS split criterion performs best in both simulations and in the analysis of real data. Our method identified two pathways that gave biological insights in the etiology of multiple myeloma. Other approaches were not able to identify any significant pathways after FDR correction, and displayed higher type I error rates in simulations. We also demonstrated that inter-marker LD does not adversely impact the prediction results for the top two pathways, or the importance measures of the top SNPs. Classification tools for GWAS may tend to choose overrepresented and large pathways, however, in our application to the multiple myeloma data set, the top two pathways are close to the first quartile of pathway size and number of SNPs.48 This suggests that our approach with FDR correction is not biased towards picking large pathways with many SNPs.
One of the advantages of our approach is that it implicitly takes into account the way SNPs may interact and it is particularly well suited for modeling pathway-based survival using SNP arrays. Pathway analysis using random forests provides a valuable tool for the researchers to combine biological information from externally available pathway databases with high-throughput data. In addition, the random forests approach provides important measures to identify SNPs that are most informative for top ranked pathways in survival prediction. These SNPs may turn out to be novel drug targets. Our approach is one of the first to combine machine learning methods with pathway information for analyzing survival SNP array data. This will greatly improve the predictive power of GWAS studies and will lead to new insights into disease mechanism.
References
Chasman DI : On the utility of gene set methods in genomewide association studies of quantitative traits. Genet Epidemiol 2008; 32: 658–668.
Peng G, Luo L, Siu H et al: Gene and pathway-based second-wave analysis of genome-wide association studies. Eur J Hum Genet 2010; 18: 111–117.
Ritchie MD : Using prior knowledge and genome-wide association to identify pathways involved in multiple sclerosis. Genome Med 2009; 1: 65.
Baranzini SE, Galwey NW, Wang J et al: Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet 2009; 18: 2078–2090.
Ballard DH, Aporntewan C, Lee JY, Lee JS, Wu Z, Zhao H : A pathway analysis applied to genetic analysis workshop 16 genome-wide rheumatoid arthritis data. BMC Proc 2009; 3 (Suppl 7): S91.
Wang K, Zhang H, Kugathasan S et al: Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet 2009; 84: 399–405.
Wang K, Li M, Bucan M : Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 2007; 81: 1278–1283.
Dinu V, Miller PL, Zhao H : Evidence for association between multiple complement pathway genes and AMD. Genet Epidemiol 2007; 31: 224–237.
Bureau A, Dupuis J, Falls K et al: Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol 2005; 28: 171–182.
Chang JS, Yeh RF, Wiencke JK et al: Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev 2008; 17: 1368–1373.
Dinu V, Zhao H, Miller P et al: Integrating domain knowledge with statistical and data mining methods for high-density genomic SNP disease association analysis. J Biomed Inform 2007; 40: 750–760.
Schumacher M, Binder H, Gerds T : Assessment of survival prediction models based on microarray data. Bioinformatics 2007; 23: 1768–1774.
van Wieringen W, Kun D, Hampel R, Boulesteix A-L : Survival prediction using gene expression data. a review and comparison. Comput Stat Data Anal 2009; 53: 1590–1603.
Pang H, Datta D, Zhao H : Pathway analysis using random forests with bivariate node-split for survival outcomes. Bioinformatics 2010; 26: 250–258.
Ishwaran H, Kogalur U, Blackstone E, Lauer M : Random survival forests. Ann Appl Stat 2008; 2: 841–860.
Hothorn T, Lausen B : On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal 2003; 43: 121–137.
Segal M : Regression trees for censored data. Biometrics 1988; 44: 35–47.
Kanehisa M, Goto S, Hattori M et al: From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 2006; 34: D354–D357.
Bender R, Augustin T, Blettner M : Generating survival times to simulate Cox proportional hazards models. Stat Med 2005; 24: 1713–1723.
International HapMap Consortium: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Browning BL, Browning SR : A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 2009; 84: 210–223.
Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A : A comprehensive evaluation of SNP genotype imputation. Hum Genet 2009; 125: 163–171.
Avet-Loiseau H, Li C, Magrangeas F et al: Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol 2009; 27: 4585–4590.
Storey JD, Tibshirani R : Statistical significance for genome-wide studies. PNAS 2003; 100: 9440–9445.
Duan S, Bleibel WK, Huang RS et al: Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res 2007; 67: 5425–5433.
Schadt EE, Molony C, Chudin E et al: Mapping the genetic architecture of gene expression in human liver. Plos Biol 2008; 6: e107.
Lauta VM : A review of the cytokine network in multiple myeloma: diagnostic, prognostic, and therapeutic implications. Cancer 2003; 97: 2440–2452.
Georgakis GV, Younes A : Cytokines and lymphomas. Cancer Treat Res 2005; 126: 69–102.
Hosgood III HD, Baris D, Zhang Y et al: Caspase polymorphisms and genetic susceptibility to multiple myeloma. Hematol Oncol 2008; 26: 148–151.
Ishitsuka K, Hideshima T, Hamasaki M et al: Honokiol overcomes conventional drug resistance in human multiple myeloma by induction of caspase-dependent and -independent apoptosis. Blood 2005; 106: 1794–1800.
Nabhan C, Gajria D, Krett NL, Gandhi V, Ghias K, Rosen ST : Caspase activation is required for gemcitabine activity in multiple myeloma cell lines. Mol Cancer Ther 2002; 1: 1221–1227.
Fuller SJ, Papaemmanuil E, McKinnon L et al: Analysis of a large multi-generational family provides insight into the genetics of chronic lymphocytic leukemia. Br J Haematol 2008; 142: 238–245.
van de Donk NW, Bloem AC, van der Spek E, Lokhorst HM : New treatment strategies for multiple myeloma by targeting BCL-2 and the mevalonate pathway. Curr Pharm Des 2006; 12: 327–340.
Lombardi L, Poretti G, Mattioli M et al: Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosomes Cancer 2007; 46: 226–238.
Carrasco DR, Tonon G, Huang Y et al: High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 2006; 9: 313–325.
Feinman R, Koury J, Thames M, Barlogie B, Epstein J, Siegel DS : Role of NF-kappaB in the rescue of multiple myeloma cells from glucocorticoid-induced apoptosis by bcl-2. Blood 1999; 93: 3044–3052.
Shi J, Tricot G, Szmania S et al: Infusion of haplo-identical killer immunoglobulin-like receptor ligand mismatched NK cells for relapsed myeloma in the setting of autologous stem cell transplantation. Br J Haematol 2008; 143: 641–653.
Tinhofer I, Marschitz I, Henn T, Egle A, Greil R : Expression of functional interleukin-15 receptor and autocrine production of interleukin-15 as mechanisms of tumor propagation in multiple myeloma. Blood 2000; 95: 610–618.
Pappa C, Miyakis S, Tsirakis G et al: Serum levels of interleukin-15 and interleukin-10 and their correlation with proliferating cell nuclear antigen in multiple myeloma. Cytokine 2007; 37: 171–175.
Alexandrakis MG, Passam FH, Sfiridaki K et al: Interleukin-18 in multiple myeloma patients: serum levels in relation to response to treatment and survival. Leuk Res 2004; 28: 259–266.
Yamashita K, Iwasaki T, Tsujimura T et al: Interleukin-18 inhibits lodging and subsequent growth of human multiple myeloma cells in the bone marrow. Oncol Rep 2002; 9: 1237–1244.
Kitano M, Ogata A, Sekiguchi M, Hamano T, Sano H : Biphasic anti-osteoclastic action of intravenous alendronate therapy in multiple myeloma bone disease. J Bone Miner Metab 2005; 23: 48–52.
Brown EE, Lan Q, Zheng T et al: Common variants in genes that mediate immunity and risk of multiple myeloma. Int J Cancer 2007; 120: 2715–2722.
Goldstein B, Hubbard A, Cutler A, Barcellos L : An application of Random Forests to a genome-wide association dataset: methodological considerations & new findings. BMC Genet 2010; 11: 49.
Genuer R, Poggi J, Tuleau C : Random forests: some methodological insights. Tech rep, INRIA 2008, http://hal.inria.fr/inria-00340725/en/, arXiv:0811.3619.
Devlin B, Risch N : A comparison of linkage disequilibrium measures for fine-mapping. Genomics 1995; 29: 311–322.
Meng Y, Yu Y, Cupples L, Farrer L, Lunetta K : Performance of random forest when SNPs are in linkage disequilibrium. BMC Bioinformatics 2009; 10: 78.
Elbers C, van Eijk K, Franke L et al: Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol 2009; 33: 419–431.
Acknowledgements
This study was supported by National Institutes of Health (grant P01CA142538) and start-up funds from Duke University Medical Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Pang, H., Hauser, M. & Minvielle, S. Pathway-based identification of SNPs predictive of survival. Eur J Hum Genet 19, 704–709 (2011). https://doi.org/10.1038/ejhg.2011.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.3
Keywords
This article is cited by
-
Random survival forests identify pathways with polymorphisms predictive of survival in KRAS mutant and KRAS wild-type metastatic colorectal cancer patients
Scientific Reports (2021)
-
Random Effects Model for Multiple Pathway Analysis with Applications to Type II Diabetes Microarray Data
Statistics in Biosciences (2015)