Abstract
Besides inducing apoptosis, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) activates NF-κB. The apoptosis signaling pathway of TRAIL is well characterized involving TRAIL receptors, Fas-associated protein with death domain (FADD) and caspase-8. In contrast, the molecular mechanism of TRAIL signaling to NF-κB remains controversial. Here, we characterized the receptor–proximal mediators of NF-κB activation by TRAIL. Deletion of the DD of TRAIL receptors 1 and 2 revealed that it is essential in NF-κB signaling. Because FADD interacts with the TRAIL receptor DD, FADD was tested. RNAi-mediated knockdown of FADD or FADD deficiency in JURKAT T-cell leukemia cells decreased or disabled NF-κB signaling by TRAIL. In contrast, TRAIL-induced activation of NF-κB was maintained upon loss of receptor interacting protein 1 (RIP1) or knockdown of FLICE-like inhibitory protein (FLIP). Exogenous expression of FADD rescued TRAIL-induced NF-κB signaling. Loss-of-function mutations of FADD within the RHDLL motif of the death effector domain, which is required for TRAIL-induced apoptosis, abrogated FADD’s ability to recruit caspase-8 and mediate NF-κB activation. Accordingly, deficiency of caspase-8 inhibited TRAIL-induced activation of NF-κB, which was rescued by wild-type caspase-8, but not by a catalytically inactive caspase-8 mutant. These data establish the mechanism of TRAIL-induced NF-κB activation involving the TRAIL receptor DD, FADD and caspase-8, but not RIP1 or FLIP. Our results show that signaling of TRAIL-induced apoptosis and NF-κB bifurcates downstream of caspase-8.
Similar content being viewed by others
Main
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a member of the TNF family, which induces several signaling pathways mediating, for example, apoptosis and activation of the transcription factor nuclear factor ‘κ-light-chain enhancer of B cells’ (NF-κB).1 Although numerous signaling steps are characterized for the TRAIL apoptosis signaling pathway,2 the molecular mechanism of NF-κB activation by TRAIL remains controversial.
Upon trimerization, TRAIL can bind to five TRAIL receptors. Of these, only TRAIL receptor 1 (TRAIL-R1, DR4) and TRAIL receptor 2 (TRAIL-R2, DR5) mediate apoptosis1 as they contain a functional death domain (DD) required for apoptosis signaling. Further DD-containing receptors of the TNF receptor family include the TNF receptor 1 (TNFR1) and cluster of differentiation 95 (CD95). To signal TRAIL-induced apoptosis, the receptor DD binds the DD-containing adaptor molecule FADD (Fas-associated protein with DD), which, in turn, recruits the initiator caspase 8 to form the death-inducing signaling complex (DISC).2
FADD and caspase-8 represent important proteins signaling TRAIL-induced apoptosis. Apart from their role in apoptosis signaling, both proteins are also known as mediators of non-apoptotic CD95 signaling or during T-cell proliferation.3, 4 FADD-deficient mice are embryonically lethal because of inadequate development of, for example, the vascular system, and FADD-deficient T cells show reduced proliferation suggesting an important role of FADD in developmental signaling.5 In the presence of receptor-interacting protein (RIP)3, caspase-8 is crucial for embryonic development as loss of caspase-8 function leads to aberrant development of the neural tube and heart.6, 7, 8
In addition to cell death, TRAIL activates survival and/or proliferation, which results, for example, in endothelial growth.9 Besides normal cells, we have shown that TRAIL activates the transcription factor NF-κB in tumor cells10 and induces survival/proliferation, which impedes the proapoptotic function of TRAIL.11, 12 The molecular mechanisms of NF-κB activation by TRAIL remain elusive, although several proteins were proposed as mediators, including TNFR-associated protein with DD (TRADD) and RIP1.13, 14
Here we aimed at characterizing the receptor–proximal signaling steps mediating TRAIL-induced activation of NF-κB using mutated TRAIL receptor fusion proteins, knockdown strategies and rescue experiments in knockout cells.
Results
LMP1-TRAIL receptor fusion proteins induce apoptosis
For signaling, TRAIL binds to five target proteins, namely TRAIL-R1–4 and osteoprotegerin. We have recently shown that TRAIL-R1/2 mediated TRAIL-induced proliferation and activation of NF-κB.11, 12 Here, we characterized the proximal signaling pathway of TRAIL-induced activation of NF-κB.
Expression constructs for TRAIL-R1 and TRAIL-R2 were generated, which induced activation of NF-κB (data not shown). Nevertheless, overexpression of wild-type receptors might lead to hetero-multimerization between recombinant and endogenous proteins disabling a clear ascription of signals to a single receptor. To circumvent this problem, fusion proteins of the TRAIL-R1/2 signaling domains with the transmembrane domain of the viral oncogene latent membrane protein 1 (LMP1) of Epstein–Barr virus were generated (Figure 1a). Signaling of such fusion proteins is specified by the fused intracellular signaling domain. Owing to the intrinsic feature to multimerize within the membrane, the LMP1 transmembrane domain renders signaling of the fusion proteins constitutively active and independent of the ligand.15 Receptor fusion proteins allowed a clear separation of signals originated by each receptor and have been successfully used before to study, for example, TNFR1 and CD40.15, 16
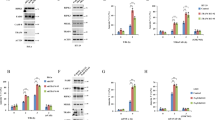
LMP1-TRAIL receptor fusion proteins induce apoptosis. (a) Scheme of different constructs used containing TRAIL receptors; numbers indicate positions of amino acids; C=C terminal region; Δ=missing; HA=hemagglutinin tag for detection; li-TEV=linker region followed by a TEV cleavage site; TM=transmembrane region; TR=TRAIL receptor. (b) The 293T cells were transfected with constructs coding for LMP1-TRAIL-R1 (LMP1-TR1) or LMP1-TRAIL-R2 (LMP1-TR2), 24 h post-transfection cells were lysed and protein expression was analyzed by western blot using the indicated antibodies; control=untransfected cells. One representative western blot of the five independent experiments is shown. (c) TRAIL apoptosis-sensitive SHEP neuroblastoma cells were transfected with plasmids coding for indicated proteins or empty vector (mock) and co-transfected with a GFP expression plasmid (ratio 4 : 1). After 24 h, percentage of living and highly GFP-expressing cells was determined by FACscan analysis upon PI staining. Percentage of surviving high GFP-expressing cells transfected with empty vector was set as 100% and used for comparison. Values represent mean of three independent experiments; error bars indicate standard error. LMP1-TR1/2=LMP1-TRAIL-R1/2
We generated a set of fusion proteins based on an HA-tagged LMP1 construct in which the LMP1 transmembrane and signaling domains were separated by a linker and a TEV protease cleavage site (J Griese and A Kieser, unpublished). The complete signaling domains of TRAIL-R1/2 and deletion mutants lacking the DDs were fused to the LMP1 transmembrane domain to generate LMP1-TRAIL-R1 and -TRAIL-R2 or LMP1-TRAIL-R1ΔC and -TRAIL-R2ΔC, respectively (Figure 1a). The negative control LMP1ΔC lacks any intracellular domain (Figure 1a).
The LMP1-TRAIL-R1/2 constructs gave rise to proteins of the expected size (Figure 1b). To confirm functionality, their proapoptotic capacity was tested in the TRAIL-sensitive neuroblastoma cell line SHEP. Overexpression of LMP1-TNFR1, LMP1-TRAIL-R1/2, but not LMP1ΔC, induced cell death (Figure 1c). Thus, receptor–fusion constructs were generated, which were functional and inducing apoptosis.
The TRAIL-R 1 and -R2 DDs are required for activation of NF-κB
To study their potential to activate NF-κB, constructs were transfected into 293T cells followed by an NF-κB luciferase reporter assay. Overexpression of the control proteins LMP1 and LMP1-TNFR1 clearly induced activation of NF-κB as did the fusion proteins LMP1-TRAIL-R1 and LMP1-TRAIL-R2 (Figure 2a). Expression of LMP1-TRAIL-R1 or LMP1-TRAIL-R2 fusion proteins was more efficient to induce NF-κB than overexpression of wildtype TRAIL-R1 or TRAIL-R2 (data not shown).
Role of FADD for TRAIL-induced activation of NF-κB – knockdown approach. (a) The 293T cells were co-transfected with the two luciferase reporter plasmids together with the constructs indicated or empty vector (mock). Luciferase activity was determined in duplicates after 24 h. Depicted is the mean of five independent experiments with standard error. LMP1-TR1/2=LMP1-TRAIL-R1/2. (b) The 293T cells were transfected twice at intervals of 24 h with siRNA directed against FADD (siFADD) or a no target sequence (control siRNA). At 24 h after last transfection, the two luciferase reporter plasmids were transfected together with LMP1-TRAIL-R2 (LMP1-TR2) or control DNA (co or control) for another 24 h. Cells were lysed and subjected to both the dual luciferase assay and western blot analysis. Luciferase assay was performed in five independent experiments measured in duplicates and is depicted as mean with standard error relative to control siRNA background level (1.0); *P<0.01 was determined by two-sided paired t-test. One representative western blot of the five experiments is shown
For TNFR1, the DD is necessary to mediate both induction of apoptosis and activation of NF-κB. An important role of the DD for activation of NF-κB was also reported for TRAIL-R2.17 To test whether activation of NF-κB by TRAIL-R1 and TRAIL-R2 depends on the receptor C terminus containing the DD in our system, fusion proteins lacking this region were used. In contrast to the fusion constructs with the entire intracellular domain of TRAIL-R1 or TRAIL-R2, LMP1-TRAIL-R1ΔC and LMP1-TRAIL-R2ΔC, as well as the non-fused variants TRAIL-R1ΔC and TRAIL-R2ΔC, were unable to activate NF-κB (Figure 2a and data not shown). These data pinpoint towards an important role of the DD for NF-κB signaling by TRAIL-R1 and TRAIL-R2. Taken together, TRAIL-R1 and TRAIL-R2 both require the C terminus containing the DD for activation of NF-κB.
FADD is essential for TRAIL-induced activation of NF-κB
Next, we aimed at identifying the receptor–proximal adaptor molecule. As activation of NF-κB by TRAIL receptor constructs depended on the presence of their C terminus containing the DD and FADD binds to the TRAIL receptors’ DDs to mediate TRAIL-induced apoptosis,2 we studied the role of FADD in TRAIL-induced activation of NF-κB.
Using a knockdown approach, the role of FADD was tested for TRAIL-induced activation of NF-κB in 293T cells. When FADD expression was transiently reduced, activation of NF-κB by LMP1-TRAIL-R2 was significantly reduced (Figure 2b). Residual LMP1-TRAIL-R2-induced NF-κB activation levels might be explained by incomplete knockdown of FADD. These data suggested a role of FADD for TRAIL-induced activation of NF-κB.
To confirm the role of FADD for TRAIL-induced activation of NF-κB in a knockout cell line, we worked with derivative variants of the T-cell leukemia cell line JURKAT. To stimulate JURKAT and derivative cells, we used TRAIL as the physiological stimulus, which also activated NF-κB in 293T cells.
We produced His- and HA-tagged soluble TRAIL, with and without N-terminal isoleucine zipper (ILZ) motif,18 as well as the functional inactive TRAIL variant C230S19 (further on named TRAIL, TRAIL C230S and ILZ-TRAIL; Supplementary Figure 1A and B), using protein isolation under denaturing conditions followed by protein refolding.20 Functional testing revealed that all TRAIL variants, except the C230S control, were functionally active and induced apoptosis and activation of NF-κB (Supplementary Figure 1C–F; see Supplementary Results for details).
Parental JURKAT cells displayed sensitivity towards TRAIL-induced apoptosis without signs of caspase-independent cell death during the time period studied (Supplementary Figure 2). TRAIL activated NF-κB via the canonical pathway with translocation of NF-κB p50 and NF-κB p65 to the nucleus as described previously (data not shown).10 In contrast, a derivative JURKAT cell line lacked the expression of FADD (JURKAT FADD def.; Figure 3a) because of chemical mutagenesis and long-term exposure to an anti-CD95 antibody. These cells are completely resistant to TRAIL-induced apoptosis.21
Role of FADD for TRAIL-induced activation of NF-κB – knockout approach and rescue. (a) Parental JURKAT and derivative JURKAT FADD def. cells were analyzed by western blot for the expression of FADD; one representative of three independent experiments is shown. (b) JURKAT and JURKAT FADD def. cells were stimulated with 1 μg/ml ILZ-TRAIL or 300 ng/ml TNFα for time periods indicated. Cytosolic lysates were analyzed for phosphorylation of IκBα, total IκBα protein and GAPDH as loading control by western blot. One representative of three independent experiments is shown. *=unspecific band detected by the α-IκBα-antibody. (c) The 293T cells were transfected with the two luciferase reporter plasmids together with an FADD expression construct or empty vector (mock). After 24 h, cells were lysed and subjected to both the dual luciferase assay and western blot. Luciferase activity was measured and is depicted relative to the empty vector (1.0); values are mean of eight independent experiments, and error bars indicate standard error. One representative western blot of the eight experiments is shown. (d) JURKAT and JURKAT FADD def. cells were transfected by nucleofection in the presence of 5 μM caspase inhibitor Q-VD-OPh with luciferase reporter plasmids and the construct coding for Flag-FADD or control DNA (co=control) as indicated. After 24 h, cells were stimulated with 1 μg/ml ILZ-TRAIL or 1 μg/ml TRAIL C230S for another 6 h. Cells were lysed and subjected to both the dual luciferase assay and western blot analysis of FADD. Luciferase activity is depicted relative to unstimulated cells (1.0); values are mean of three independent experiments measured in duplicates, and error bars indicate standard error. One representative western blot of the three independent experiments is shown
Importantly and in contrast to parental JURKAT cells, FADD-deficient JURKAT cells were completely unable to activate NF-κB upon stimulation with TRAIL as shown by western blot analysis of phosphorylated inhibitor of the κB protein α (IκBα) (Figure 3b), luciferase reporter assay and enzyme-linked immunosorbent assay (ELISA) detecting activated NF-κB p65 (data not shown).
Thus, two independent cellular systems with low or absent expression of FADD showed reduced activation of NF-κB by TRAIL using either soluble TRAIL or TRAIL receptor constructs as stimulants. These data strongly suggested that the adaptor molecule FADD mediated TRAIL-induced activation of NF-κB.
Rescue by overexpression of recombinant FADD
To prove that FADD deficiency is responsible for loss of TRAIL-induced activation of NF-κB in FADD-deficient JURKAT cells, a rescue experiment was performed, as they might have acquired further alterations in addition to loss of FADD expression. The transient overexpression of Flag-FADD22 in 293T cells alone induced marked activation of NF-κB in the absence of TRAIL (Figure 3c).
In FADD-deficient JURKAT cells, transient expression of FADD clearly rescued the ability of soluble TRAIL to activate NF-κB (Figure 3d). As a conclusion, the loss of FADD in JURKAT FADD def. cells disabled TRAIL-induced activation of NF-κB. Our data prove that FADD represents the adaptor protein mediating TRAIL-induced activation of NF-κB in JURKAT cells.
Critical role of the 182RHDLL186 motif of FADD for TRAIL-induced apoptosis
The highly conserved motif RHDLL within the death effector domain (DED) of FADD is crucial for induction of apoptosis by TRAIL.23 In CD95 signaling, mutations within the RHDLL motif inhibit FADD multimerization, apoptosis signaling downstream of FADD and recruitment of caspase-8, despite effective binding of FADD to the receptor. The same mutations within the RHDLL motif abrogate apoptosis induction by TRAIL.23 We therefore asked for the role of this motif within TRAIL-induced activation of NF-κB.
To avoid effects of endogenous wild-type FADD protein, experiments were performed in FADD-deficient JURKAT cells, which were transduced using lentiviruses. As FADD is a proapoptotic protein, an inducible expression system was applied to avoid premature cell death. Inducible transgene expression was based on post-translational regulation mediated by fusion to an FKBP (FK506 binding protein)-based domain targeting the protein to rapid proteosomal degradation. Addition of the synthetic ligand Shield-1 allows inducible protein stabilization and enables cellular function of the recombinant protein.24 As the FKBP domain is about 12 kDa in size, the fusion proteins were accordingly larger in western blot compared with the endogenous protein.
Similar to the work carried out by Muppidi et al.,23 we cloned two derivatives of FADD, each containing two point mutations within the RHDLL motif, namely the AHALL mutant containing the R182A and D184A mutations, and the RHDAA mutant (the underline emphasizes the mutated amino acids) containing the L185A and L186A mutations (Figure 4a). The expression of wild-type FKBP-FADD and the two mutants was inducible in a dose-dependent manner, although both mutants showed leakiness of expression in the absence of Shield-1 (Figure 4b and Supplementary Figure 3). As expected, parental JURKAT cells were sensitive and FADD-deficient cells were resistant towards TRAIL-induced apoptosis (Figure 4c, left panel). Expression of wild-type FKBP-FADD in FADD-deficient cells rescued their sensitivity towards TRAIL-induced apoptosis. Higher FKBP-FADD protein levels correlated with higher levels of apoptosis induction (Figure 4c, right panel). In contrast, overexpression of either of the two mutants carrying the AHALL or RHDAA motif did not result in apoptosis induction, although the proteins were expressed in supra-physiological levels (Figure 4c, right panel and Supplementary Figure 3B). Accordingly, reconstitution of FADD-deficient cells with FKBP-FADD, but neither FKBP-AHALL nor FKBP-RHDAA, restored cleavage of caspase-8, FLIPL (FLICE-like inhibitory protein long) or RIP1 (Figure 4d).
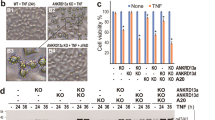
Role of the 182RHDLL186 motif of FADD for TRAIL-induced apoptosis. (a) Scheme of lentiviral constructs for inducible expression of FADD and its mutants; numbers indicate positions of amino acids; letters indicate amino acids according to the conventional abbreviation; FKBP=degradation domain; wt=wild type. (b) JURKAT FADD def. cells stably inheriting the wt FKBD-FADD expression construct were incubated with Shield-1 at concentrations indicated for 72 h. Western blot shows expression levels of the wild-type FKBP-FADD protein in JURKAT FADD def. cells in comparison to endogenous FADD in parental JURKAT cells; mock=empty vector; one representative of two independent experiments is shown. (c) Stably transduced parental JURKAT cells or JURKAT FADD def. cells carrying different expression constructs or empty vector (mock) were stimulated with ILZ-TRAIL at concentrations indicated for 48 h; apoptosis was measured by forwardside scatter analysis. Values are representative of two independent experiments measured in duplicates. Left panel shows controls, and right panel shows JURKAT FADD def. derivative cell lines subjected to Shield-1 for 24 h at concentrations indicated to induce expression of the different transgenes. Expression of transgenes in this experiment is shown in panel b and Supplementary Figure 3B. (d) A total of 1 × 106 JURKAT FADD def. cells reconstituted with the indicated FADD variants were pretreated for 24 h with 0.06 μM Shield-1 (or 0.5 μM for mutants) and stimulated with ILZ-TRAIL (1 μg/ml) for 1 or 2 h. Processing of caspase-8, RIP1 and FLIPL in whole cell lysates was analyzed by western blot. *=unspecific band; GAPDH served as loading control
To study the role of the RHDLL motif for DISC formation, we performed co-immunoprecipitation experiments in FADD-deficient JURKAT cells stably reconstituted with either FKBP-FADD or FKBP-AHALL. Although both wild-type and mutant FADD were able to bind to the TRAIL receptors, only FKBP-FADD, but not FKBP-AHALL, was able to recruit caspase-8 to the DISC (Figure 5). These data suggest that the 182RHDLL186 motif of FADD mediates recruitment of caspase-8 upon stimulation with TRAIL.
DISC recruitment in reconstituted JURKAT cells. A total of 5 × 107 JURKAT FADD def. cells reconstituted with the indicated FADD variants or empty vector (mock) were pretreated with 0.06 μM Shield-1 for 24 h (except mock) and stimulated or not with 15 μg ILZ-TRAIL for 1 h. Cells were lysed and immunoprecipitation was performed using anti-HA-coupled beads precipitating HA-TRAIL; lysates and immunoprecipitates were analyzed by western blot for the presence of TRAIL-R2, FADD and caspase-8. GAPDH served as loading control; one representative out of two independent experiments is shown. *=unspecific band
The 182RHDLL186 motif of FADD is essential for TRAIL-induced activation of NF-κB
Next, we tested the different FKBP-FADD constructs with regard to their influence on TRAIL-induced activation of NF-κB in FADD-deficient JURKAT cells. In accordance to the data obtained in transient transfection (Figure 3d), FKBP-FADD rescued activation of NF-κB by TRAIL in FADD-deficient JURKAT cells. NF-κB activation was detected by phosphorylation of IκBα and binding of activated p65 to DNA (Figures 6a and b). Thus, both transient (Figure 3d) and stable re-expression of FADD (Figures 6a and b) rescued TRAIL-induced activation of NF-κB.
Role of the 182RHDLL186 motif of FADD for TRAIL-induced activation of NF-κB. (a) JURKAT FADD def. wild-type FKBP-FADD cells were stimulated with 0.5 μM Shield-1 for 24 h and thereafter with either TNFα (300 ng/ml) or ILZ-TRAIL (1 μg/ml) for the time periods indicated. Lysates were analyzed by western blot as in Figure 3b; one representative of two independent experiments is shown. (b, c) JURKAT cells or JURKAT FADD def. cells stably transduced with the different expression cassettes were subjected to 0.5 μM Shield-1 (except wild-type FKBP-FADD: 0.06 μM) for 24 h. Thereafter, 1 μg/ml ILZ-TRAIL was added for the time periods indicated and NF-κB activity was determined by DNA binding of activated p65 detected using ELISA. Values are representative of three (b) or two (c) independent experiments measured in triplets. Expression of transgenes in these experiments is shown in Supplementary Figure 4
In contrast and despite supra-physiological expression levels of mutant proteins (Supplementary Figure 4), both the AHALL and RHDAA mutants were unable to rescue TRAIL-induced activation of NF-κB (Figure 6c). These data show that the pentameric 182RHDLL186 motif of FADD is necessary for the activation of NF-κB by TRAIL suggesting the requirement of caspase-8 recruitment. Taken together, FADD mediated TRAIL-induced activation of NF-κB depended on the 182RHDLL186 motif within its DED, which is also required for TRAIL-induced apoptosis.
RIP1 and FLIP are not essential for TRAIL-induced activation of NF-κB
RIP1 was suggested to be essential for NF-κB activation by TRAIL in MEFs13 and represents another adaptor molecule binding to death receptors independently of FADD.25 To study the role of RIP1 for TRAIL-induced activation of NF-κB, DISC immunoprecipitations were performed in FADD-deficient JURKAT cells (Figure 7a). RIP1 was clearly detectable at the DISC in the absence of FADD, while reconstitution with FADD abrogated RIP1 recruitment to the DISC suggesting competitive binding of FADD and RIP1 to TRAIL receptors. Furthermore, recruitment of RIP1 to the DISC in FADD-deficient cells was not sufficient to activate NF-κB (Figure 6b).
RIP1 is not required for TRAIL-induced activation of NF-κB in JURKAT cells. (a) Same experiment as in Figure 5, this time visualizing RIP1 binding to the DISC. TRAIL-R2 is shown equally as in Figure 5, as it serves as control for comparison. (b) A total of 2 × 106 parental JURKAT cells or JURKAT RIP def. cells were stimulated or not with 1 μg/ml ILZ-TRAIL for 1 h and expression of RIP1 and processing of caspase-8 was analyzed by western blot in whole cell lysates. (c) Parental JURKAT cells or JURKAT RIP def. cells were stimulated with ILZ-TRAIL at concentrations indicated for 48 h; apoptosis values are representative of two independent experiments measured in duplicates. (d) JURKAT cells or JURKAT RIP def. cells were stimulated with 300 ng/ml TNFα or 1 μg/ml ILZ-TRAIL for the indicated time periods and NF-κB activity was determined by DNA-binding of activated p65 detected by ELISA. Values are representative of two independent experiments measured in duplicates
To further validate the role of RIP1 for NF-κB activation by TRAIL, a RIP-deficient JURKAT cell line was tested.26 RIP1 deficiency had no influence on processing of caspase-8 and induction of apoptosis by TRAIL (Figures 7b and c). In contrast to TNFα, TRAIL activated NF-κB in RIP1-deficient cells to a similar extent as in parental cells (Figure 7d). Thus, RIP1 was neither sufficient nor necessary for TRAIL-induced activation of NF-κB.
Similarly, knockdown of FLIP did not inhibit TRAIL-induced activation of NF-κB; NF-κB activation by LMP1-TRAIL-R2 in 293T cells was rather increased upon stable or transient knockdown of FLIP (Supplementary Figure 5). These data are in line with published results showing an inhibitory function of FLIP for TRAIL-induced activation of NF-κB.27, 28
Caspase-8 is essential for TRAIL-induced activation of NF-κB
In CD95-induced apoptosis, recruitment of caspase-8 to FADD depends on the RHDLL motif of FADD.23 Here, TRAIL-induced apoptosis and activation of NF-κB depended on the RHDLL motif of FADD. Therefore, we asked whether caspase-8 represents the next step of TRAIL-induced activation of NF-κB.
Stable or transient knockdown of caspase-8 in 293T cells significantly decreased levels of NF-κB activation induced by overexpression of LMP1-TRAIL-R2 (Figures 8a and b and data not shown). These data suggested a potential role of caspase-8 for TRAIL-induced activation of NF-κB. In addition, overexpression of caspase-8 alone strongly induced NF-κB in 293T cells (Supplementary Figure 6A).
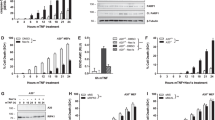
Role of caspase-8 for TRAIL-induced activation of NF-κB. (a) The 293T cells were stably transduced with empty vector (293T mock) or lentiviral particles containing shRNA directed against caspase-8 (293T shC8). Cells were transfected with luciferase reporter plasmids together with LMP1-TR2 or control DNA (co). Luciferase activity was determined after 24 h and is depicted relative to background level (1.0) of cells transfected with control DNA. Values represent mean of 20 independent experiments; error bars indicate standard error. *P<0.01 determined by two-sided paired t-test. LMP1-TR2=LMP1-TRAIL-R2. (b) Expression of caspase-8 in 293T mock and 293T shC8 cells (see a) was as detected by western blot; one representative of two independent experiments is shown. (c) Expression of caspase-8 in JURKAT and JURKAT C8 def. cells as detected by western blot; one representative of two independent experiments is shown. (d) JURKAT C8 def. cells were stimulated with either TNFα (300 ng/ml) or ILZ-TRAIL (1 μg/ml) for time periods indicated; cytosolic lysates were analyzed for phosphorylation of IκBα, total IκBα protein and GAPDH as loading control by western blot; one representative of three independent experiments is shown. (e) Parental JURKAT and JURKAT caspase-8 def. cells reconstituted with caspase-8 or empty vector (mock) were stimulated for 1 or 2 h with 0.5 μg/ml ILZ-TRAIL and analyzed by western blot for the expression of caspase-8. GAPDH served as loading control. (f) A total of 5 × 106 JURKAT or JURKAT caspase-8 def. cells reconstituted with caspase-8 or empty vector (mock) were stimulated with 2.5 μg ILZ-TRAIL for the indicated time periods and NF-κB activity was determined by DNA binding of activated p65 detected using ELISA. Values are representative of four independent experiments measured in duplicates. (g) Parental JURKAT and JURKAT caspase-8 def. cells reconstituted with caspase-8-FKPB, its catalytically inactive mutant C360S or empty vector (mock) were analyzed by western blot for the expression of endogenous caspase-8 (55 kDa) and caspase-8-FKBP (65 kDa) or mutant caspase-8 C360S-FKBP (65 kDa). GAPDH served as loading control. (h) A total of 2 × 106 stably transduced parental JURKAT cells or JURKAT C8 def. cells carrying the different expression constructs or empty vector (mock) were stimulated for the indicated periods of time with 2.5 μg ILZ-TRAIL. Cells expressing FKBP-tagged proteins were pretreated with 0.5 μM Shield-1 for 24 h. Whole cell extracts were prepared and NF-κB activity was determined by DNA binding of activated p65 detected using ELISA
To confirm a putative role of caspase-8 for TRAIL-induced activation of NF-κB, a derivative cell line of JURKAT was chosen which lacks expression of caspase-8,29 resulting in resistance towards TRAIL-induced apoptosis (Figure 8c and data not shown).
Stimulation of caspase-8-deficient cells with ILZ-TRAIL was not able to induce phosphorylation of IκBα as readout for activation of NF-κB, whereas TNFα clearly did so (Figure 8d). Similar results were obtained using the luciferase reporter assay as readout and ELISA detecting activated NF-κB p65 protein (Figure 8f and Supplementary Figure 6B). These data suggested a role of caspase-8 for TRAIL-induced activation of NF-κB.
The active site of caspase-8 is essential for TRAIL-induced activation of NF-κB
To ensure that loss of caspase-8 was responsible for loss of NF-κB activation by TRAIL, rescue experiments were performed. Recombinant caspase-8 was stably expressed in caspase-8-deficient cells and thereby rescued activation of NF-κB by TRAIL in these cells, despite rather low expression levels of exogenous caspase-8 (Figures 8e and f). Thus, caspase-8 is essential for TRAIL-induced activation of NF-κB in JURKAT cells.
We next asked whether the active site 358QACQG362 of caspase-8 is required for TRAIL-induced activation of NF-κB. This remains unclear, as caspase inhibition, for example, by z-VAD allows initial processing of caspase-8 and cleavage of FLIPL, as well as TRAIL-induced activation of NF-κB (data not shown).30 For these experiments, we used our FKBP-based inducible expression system, but protein expression remained leaky and non-inducible (data not shown).
In parallel to the expression of caspase-8, caspase-8-FKBP protein levels remained very low, while mutant caspase-8-C360S-FKBP was expressed at physiological levels (Figure 8g). Nevertheless, low levels of wild-type, but not high levels of mutant, caspase-8 rescued TRAIL-induced activation of NF-κB in caspase-8-deficient JURKAT cells (Figure 8h). These data show that the active site of caspase-8 is required for TRAIL-induced activation of NF-κB.
Taken together, the adaptor molecule FADD with its RHDLL motif and the initiator caspase-8 with its active site, but neither RIP1 nor FLIP, mediate TRAIL-induced activation of NF-κB.
Discussion
Our study shows that TRAIL-induced activation of NF-κB depends on the C terminus of TRAIL-R1 and TRAIL-R2 containing the DD, on FADD and the RHDLL motif of FADD, as well as on caspase-8 and its active site. These conclusions are based on (i) stimulating the signaling pathway either by soluble TRAIL or by transfection of LMP1-TRAIL receptor fusion proteins, (ii) the use of either small interfering RNA (siRNA)-mediated knockdown or expression-deficient cell lines and (iii) rescue experiments also using different mutants of FADD and caspase-8. Involvement of FADD and caspase-8 thus represent essential steps of both pathways activated by TRAIL, apoptosis and activation of NF-κB.
Both receptors, TRAIL-R1 and TRAIL-R2, were dependent on their DD to induce NF-κB as well as apoptosis; accordingly, both LMP1 fusion proteins of TRAIL-R1 and TRAIL-R2 preventing potential interaction with endogenous receptors induced activation of NF-κB only as long as their DD was present. These data suggest that both receptors might induce identical downstream signaling steps.
At the level of the TRAIL DISC, various further proteins were discussed as mediators of TRAIL-induced activation of NF-κB. The role of TRADD is controversially discussed.31 Endogenous TRADD was not reliably detected at the native TRAIL-DISC.32 Nevertheless, influence of TRADD on TRAIL-induced activation of NF-κB was reported.14, 33 Our data do not favor a role of TRADD for TRAIL-induced activation of NF-κB, as FADD-deficient JURKAT cells express TRADD, but lack activation of NF-κB by TRAIL.
For activation of NF-κB, TNFα via TNFR1 recruits TRADD, TRAF2 and RIP.34 We showed here that TRAIL-R1/2 recruited FADD and caspase-8 for activation of NF-κB independent of RIP1. Thus, clear differences exist between signaling by TNFR135 and TRAIL-R1/2, while TRAIL-R1/2 signaling shares high similarities to CD95 signaling.3
In the CD95 DISC, caspase-8 is recruited to FADD dependent on the RHDLL motif.23 We now showed that the RHDAA and AHALL variant of FADD was not able to rescue TRAIL-induced activation of NF-κB in FADD-deficient cells and that recruitment of caspase-8 to the TRAIL DISC was required for downstream signaling.
Our results show that the active site of caspase-8 is required for signaling TRAIL-induced activation of NF-κB. In line with this, Choi et al.36 observed caspase-dependent induction of the NF-κB target interleukin-8 by TRAIL. Nevertheless, Harper et al.37 described increased NF-κB activation by caspase inhibition via z-VAD-fmk, and Wajant et al.38 found NF-κB activation by TRAIL to be independent of caspase inhibition.
Studies on heterozygous caspase-8 knockout mice overexpressing a catalytically inactive caspase-8 showed increased inflammation, although the role of NF-κB for the pathogenesis remains unclear.39 In contrast to our cells which completely lack expression of caspase-8, these mice retain caspase-8 function by one allele.
Downstream of the DISC, in CD95 signaling a direct interaction between the p43 cleavage product of FLIPL and members of the IκB kinase (IKK) complex was described, which depended on the presence and catalytic activity of caspase-8.30 A role of RIP1 for mediating NF-κB activation by CD95 has also been reported,3 while NF-κB activation by TRAIL was independent of RIP1 in our hands. We detected cleaved p43 FLIPL upon stimulation with TRAIL in cells expressing FADD. In contrast, knockdown of FLIP did not inhibit, but even slightly, increased activation of NF-κB by TRAIL, as it has been described before.28 This is in line with the reported inhibitory effect of high levels of full-length FLIPL on NF-κB activation by TRAIL.27 Another model for a potential signaling of TRAIL-induced activation of NF-κB downstream of the DISC represents the formation of a proposed secondary signaling complex independent of TRAIL-R1/2, which recruits proteins like IKKα and NF-κB essential modulator.32, 40
During anticancer treatment with TRAIL, it would be advantageous to activate selectively TRAIL-induced apoptosis without activation of NF-κB. Here we show that receptor–proximal signaling is identical between both pathways. Studies of further downstream signaling steps are therefore necessary to define potential targets for inhibition of TRAIL-induced signaling of survival and proliferation during anticancer therapy. Furthermore, TRAIL activates additional prosurvival pathways like c-Jun N-terminal kinase and mitogen-activated protein kinase signaling, which might participate in mediating TRAIL-induced proliferation of tumor cells besides activation of NF-κB.12 It remains to be determined whether inhibition of these pathways in addition to inhibition of NF-κB might be beneficial during anticancer therapy.
Taken together, we describe that TRAIL activates NF-κB by the DDs of TRAIL-R1 and TRAIL-R2. FADD and caspase-8 represent the adaptor molecule and proximal signaling protein mediating TRAIL-induced activation of NF-κB independent of RIP1 and FLIP. Thus, TRAIL-induced apoptosis and NF-κB signaling bifurcates downstream of caspase-8.
Materials and Methods
Reagents and cell lines
Caspase inhibitor Q-VD-OPh was purchased from Merck (Darmstadt, Germany), and recombinant human TNFα and recombinant human TRAIL from PeproTech (Hamburg, Germany). Shield-1 was obtained from Cheminpharma LCC (Farmington, CT, USA). All other reagents were purchased from Sigma-Aldrich (Hannover, Germany) or Carl Roth (Karlsruhe, Germany). For detection in immunoblotting, the following antibodies were used: α-tubulin and α-GAPDH (Merck), α-caspase 8 (Enzo Life Sciences, Farmingdale, NY, USA), caspase-8 (N-term) rabbit (Epitomics Inc., Burlingame, CA, USA), α-FADD (BD Biosciences, Franklin Lakes, NJ, USA), α-FADD rabbit (Epitomics Inc.), α-p-IκBα (Cell Signaling, Danvers, MA, USA), α-FLIP (Enzo Life Sciences), α-IκBα (Santa Cruz Technology, Santa Cruz, CA, USA), α-RIP (BD Biosciences), α-RIP rabbit(Cell Signaling) α-TRAIL-R1 (ProSci, Poway, CA, USA) and α-TRAIL-R2 (Abcam, Cambridge, UK). The α-HA mAb (3F10) was kindly provided by Elisabeth Kremmer (Munich, Germany). As secondary antibodies, α-mouse IgG1 (Southern Biotech, Birmingham, AL, USA), α-mouse IgG and α-rabbit IgG (Cell Signaling), as well as α-rat (Merck) were used, all coupled to horseradish peroxidase. JURKAT (A3) cells, JURKAT FADD def. and JURKAT caspase-8 def. were kindly provided by John Blenis (Boston, MA, USA). RIP-deficient JURKAT cells were a kind gift from Brian Seed (Boston, MA, USA). All other cell lines were obtained from DSMZ (Heidelberg, Germany). Cells were cultured in RPMI, or in case of 293T cells in DMEM (Life Technologies GmbH, Darmstadt, Germany), all were supplemented with 10% fetal calf serum (Biochrom AG, Berlin, Germany) and 1% glutamine (Life Technologies GmbH).
Constructs, cloning, PCR mutagenesis and siRNA
Constructs were generated in pET17b (Merck), pGreen Puro and pCDH (System Bioscience, Mountain View, CA, USA), as well as pCMV.15 Templates for human TRAIL, TRAIL-R1 and TRAIL-R2 were obtained as cDNA clones from imaGenes GmbH (Berlin, Germany); the coding sequence for the ILZ and tags, flanked with restriction sites NdeI and BamHI, was synthesized at GeneArt (Regensburg, Germany). To clone TRAIL variants, the coding sequence of soluble TRAIL was inserted into pET17b between BamHI and EcoRI restriction sites. The synthesized DNA was inserted in front of the coding sequence of TRAIL using present NdeI and BamHI sites. Different parts of the synthesized DNA were subsequently removed with the help of pairwise flanking restriction sites to generate the different TRAIL variants. To clone TRAIL receptor fusion proteins, the intracellular parts of the TRAIL receptors (for details see Figure 1a) were amplified by PCR and inserted between AflII and BlpI sites into pCMV vector containing LMP1, thereby replacing the signaling domain of LMP1. For stable expression of FADD, the lentiviral vector pCDH containing a GFP expression cassette was used. The Flag-FADD expression vector,22 also used for transient experiments, served as template for subcloning of FADD. To allow inducible protein expression, an FKBP(L106P) domain24 was inserted between XbaI and EcoRI sites (for N-terminal fusion) or at the NotI site (for C-terminal fusion) of pCDH. The coding sequence of FADD and caspase-8 was inserted between EcoRI and NotI sites of vectors with N-terminal FKBP domain for FADD and C-terminal FKBP domain for caspase-8: the stop codons were removed by PCR to allow transcription of one open reading frame coding for FADD/caspase-8 and GFP, connected by a ‘self-cleaving’ T2A peptide. In addition, a puromycin cassette derived from vector pGreen Puro was amplified with a T2A sequence at the beginning and no stop codon at the end, and introduced at the NotI site to allow selection of transduced cells. The construct pGreen shcaspase-8 and FLIP was generated according to the manufacturer’s instructions based on the siRNA sequence for caspase-8 (5′-GGGTCATGCTCTATCAGAT-3′) or for FLIP (5′-CTACGAATAGCCACTGCATACCAAC-3′). To generate TRAIL C230S, the truncated death receptor proteins, the FADD mutants AHALL and RHDAA, as well as caspase-8 C360S PCR mutagenesis was performed using QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (Santa Clara, CA, USA). Plasmids of all constructs were confirmed by sequencing. All used oligonucleotides and siFADD (5′-ACGTCATATGTGATAATGT-3′) were synthesized at Eurofins MWG Operon (Ebersberg, Germany).
TRAIL production
Detailed procedures are described in Supplementary Methods. Briefly, protein was expressed in Escherichia coli BL21(DE3)9 and extracted under denaturing conditions. Activity of TRAIL and TRAIL variants was restored by stepwise dilution with refolding buffer. For storage, TRAIL was dialyzed to storage buffer. Concentration determination was carried out by comparison with BSA standard in a Coomassie-stained SDS gel. LPS content of a TRAIL preparation was <1 ng/μg protein, determined by LAL-Assay at Profos AG (Regensburg, Germany).
Transient transfection
The 293T and SHEP cells were transiently transfected with DNA or siRNA using TurboFect in vitro Transfection Reagent (Fermentas, St. Leon-Rot, Germany) according to the manufacturer’s instructions. JURKAT cell lines were transiently transfected using the Amaxa Human T Cell Nucleofector Kit (Lonza Group Ltd, Basel, Switzerland) following the manufacturer’s instructions.
Production of lentiviral particles, transduction, generation of stable cell lines and proteo tuning
The production of lentiviral particles is described in detail in Supplementary Methods. For transduction, cells were transduced overnight with lentiviral particles in the presence of 3 μg/ml Polybren (Sigma-Aldrich) in culture medium as described above. The next day, cells were washed in phosphate-buffered saline (PBS) and cultured in fresh medium. For stable knockdown of caspase-8 and FLIP, 293T cells were infected twice with a time lag of 3 weeks with lentiviral particles derived from lentivector pGreen shcaspase-8 or shFLIP and selected after 24 h with puromycin (10 μg/ml for 24 h). Knockdown efficiency was verified by western blot analysis of caspase-8 or FLIP expression levels. Transduction of JURKAT cell lines with lentiviral particles produced with pCDH-derived lentivectors was performed once, followed 24 h later by selection with puromycin (5 μg/ml for 72 h) or FACS sorting. Efficiency of transduction was determined by analysis of GFP expression. To induce FKBP-modulated transgene expression, Shield-1 at a final concentration of 0.5 μM (or 0.06 μM for FKBP-FADD) was added to the stably transduced cell lines for 24 h if not stated differently and protein expression was monitored by western blot analysis.
Luciferase assay
For each condition, 105 293T cells or 106 cells of JURKAT cell lines were transfected with reporter plasmids 3x-κB-L (5 ng for 293T, 1.7 μg for JURKAT)15 and phosphoglycerate kinase promoter-driven Renilla control reporter (25 ng for 293T, 0.7 μg for JURKAT), as well as 970 ng of salmon sperm DNA for 293T cells or 2.7 μg Flag-FADD vector or salmon sperm DNA for JURKAT cells. Cells were stimulated 24 h later with TRAIL variants (1 μg/ml) or TNFα (300 ng/ml) for another 6 h and lysed with passive lysis buffer according to the manufacturer’s instructions. Luciferase activity was determined by Dual Luciferase Assay (Promega, Madison, WI, USA) and values of firefly luciferase were normalized to values of Renilla luciferase. Levels of unstimulated cells were set as 1.0.
To measure NF-κB activation by the expression of TRAIL receptor fusion proteins, 293T cells were transfected with reporter plasmids 3x-κB-L (5 ng) and PGK Renilla (25 ng), as well as 500 ng of the vector coding for the fusion protein of interest and 470 ng of salmon sperm DNA. To investigate the influence of transient knockdown of FADD or FLIP on NF-κB activation by LMP1-TRAIL-R2, 293T cells were transfected twice with 150 pmol of small interfering RNA (siRNA) 24 and 48 h before transfection with reporter and expression plasmids. In case of stable knockdown of caspase-8 or FLIP, 239T cells were stably transduced with small hairpin RNA (shRNA) against caspase-8/FLIP or empty vector (293T shC8, 293T mock) were transiently transfected with reporter and expression vectors. At 24 h after transfection of the reporter and expression vectors, luciferase assay was performed. Levels of cells transduced with empty vector were set as 1.0.
Apoptosis analysis
To measure apoptosis induction by TRAIL variants, cells were stimulated for 48 h and apoptosis was measured by forward side scatter analysis controlled by Annexin V/propidium iodide staining in FACS Calibur (BD Biosciences). For specific inhibition of TRAIL by TRAIL-R2(hu):Fc(hu) (Enzo Life Sciences), TRAIL was preincubated for 1 h at 4°C with 10-fold excess of TRAIL-R2(hu):Fc(hu) before stimulation of cells for 48 h.
For Annexin V/ethidium homodimer III staining, 1 × 106 cells were preincubated for 24 h with or without 5 μM Q-VD-OPh and subsequently stimulated with 1 μg ILZ-TRAIL for 2 or 6 h. The staining was performed using the Apoptosis, Necrosis and Healthy Cell Quantitation Kit Plus (Biotium, Hayward, CA, USA) according to the manufacturer’s instructions. Fluorescence was measured with the LSRFortessa cell analyzer (BD Biosciences).
A total of 5 × 104 SHEP cells were transfected with 800 ng of the respective constructs together with 200 ng of a vector coding for GFP. After 24 h, the survival rate was determined by gating on highly GFP-expressing cells.
Cell lysis and western blot analysis
To validate protein expression, whole cell lysates were prepared by lysis of 106 cells in 100 μl lysis buffer (43 mM Tris, 9% glycerol, 1.8% SDS, 4.5% β-mercaptoethanol, 0.05% bromophenol blue); otherwise, lysates from Dual Luciferase Assays and the TransAM p65 ELISA (Active Motif, Rixensart, Belgium) were used. For detection of phosphorylated IκBα, cells were stimulated with ILZ-TRAIL (1 μg/ml) or TNFα (300 ng/ml) for the indicated periods and lysed for 20 min with 100 μl cytoplasmic lysis buffer (10 mM HEPES; 1 mM KCl; 1.5 mM MgCl2; 0.5% Triton X-100 (pH 7.0)) supplemented with protease inhibitor cocktail (Merck) and phosphatase inhibitor PhosSTOP (Roche, Basel, Switzerland).
For analysis, lysates were separated on 12% SDS-polyacrylamide gels and transferred to PVDF membranes. Membranes were hybridized with specific antibodies for GAPDH or α-tubulin as loading controls or antibodies specific for caspase-8, FADD, TRAIL-R1, TRAIL-R2, p-IκBα and IκBα, as well as the appropriate secondary antibodies. For detection SuperSignal West Femto Chemiluminescent Substrate (Thermo Fisher, Rockford, IL USA) was used.
Co-immunoprecipitation
A total of 5 × 107 cells were incubated with or without ILZ-TRAIL (15 μg) for 1 h, washed with ice-cold PBS and lysed with 1 ml of buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, pH 8.0). Co-immunoprecipitation was performed using the Pierce HA Tag IP/Co-IP Kit (Thermo Fisher) according to the manufacturer’s instructions and subsequently analyzed by western blot analysis. For the detection of FADD and caspase-8 in immunoprecipitates, monoclonal rabbit antibodies from Epitomics Inc. were used. RIP1 was detected with monoclonal rabbit antibody from Cell Signaling.
NF-κB binding activity
A total of 2 × 106 (or 5 × 106) cells per time point were stimulated with 1 μg ILZ-TRAIL (or 2.5 μg) for the indicated periods. Whole cell lysates were prepared and used directly for the DNA-binding assay using reagents of the TransAM p65 ELISA Kit according to the manufacturer’s instructions. Wells were developed for 10 min and absorbance measured at a wavelength of 450 nm. Basal level of unstimulated cells was set as 1.0.
Statistical analysis
Statistical analysis was performed by two-sided paired t-test. P’s<0.05 were considered significant. Data were presented as mean of at least five independent experiments with standard error.
Abbreviations
- CD95:
-
cluster of differentiation 95
- DD:
-
death domain
- DED:
-
death effector domain
- DISC:
-
death inducing signaling complex
- ELISA:
-
enzyme-linked immunosorbent assay
- FADD:
-
Fas-associated protein with death domain
- FLIPL:
-
FLICE-like inhibitory protein long
- FKBP:
-
FK506 binding protein
- IκBα:
-
inhibitor of the κB protein α
- IKK:
-
IκB kinase
- ILZ:
-
isoleucine zipper
- LMP1:
-
latent membrane protein 1
- NF-κB:
-
nuclear factor ‘κ-light-chain enhancer of B cells’
- RIP:
-
receptor-interacting protein
- siRNA:
-
small interfering RNA
- shRNA:
-
small hairpin RNA
- TNF:
-
tumor necrosis factor
- TNFR1:
-
TNF receptor 1
- TRADD:
-
TNFR-associated protein with death domain
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- TRAIL-R1/2:
-
TRAIL receptor 1/2
References
Schneider P, Thome M, Burns K, Bodmer JL, Hofmann K, Kataoka T et al. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity 1997; 7: 831–836.
Gonzalvez F, Ashkenazi A . New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 2010; 29: 4752–4765.
Kreuz S, Siegmund D, Rumpf JJ, Samel D, Leverkus M, Janssen O et al. NFkappaB activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. J Cell Biol 2004; 166: 369–380.
Pellegrini M, Bath S, Marsden VS, Huang DC, Metcalf D, Harris AW et al. FADD and caspase-8 are required for cytokine-induced proliferation of hemopoietic progenitor cells. Blood 2005; 106: 1581–1589.
Yeh WC, Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A et al. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 1998; 279: 1954–1958.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Sakamaki K, Inoue T, Asano M, Sudo K, Kazama H, Sakagami J et al. Ex vivo whole-embryo culture of caspase-8-deficient embryos normalize their aberrant phenotypes in the developing neural tube and heart. Cell Death Differ 2002; 9: 1196–1206.
Secchiero P, Gonelli A, Carnevale E, Milani D, Pandolfi A, Zella D et al. TRAIL promotes the survival and proliferation of primary human vascular endothelial cells by activating the Akt and ERK pathways. Circulation 2003; 107: 2250–2256.
Jeremias I, Kupatt C, Baumann B, Herr I, Wirth T, Debatin KM . Inhibition of nuclear factor kappaB activation attenuates apoptosis resistance in lymphoid cells. Blood 1998; 91: 4624–4631.
Ehrhardt H, Fulda S, Schmid I, Hiscott J, Debatin KM, Jeremias I . TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene 2003; 22: 3842–3852.
Baader E, Toloczko A, Fuchs U, Schmid I, Beltinger C, Ehrhardt H et al. Tumor necrosis factor-related apoptosis-inducing ligand-mediated proliferation of tumor cells with receptor-proximal apoptosis defects. Cancer Res 2005; 65: 7888–7895.
Lin Y, Devin A, Cook A, Keane MM, Kelliher M, Lipkowitz S et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol Cell Biol 2000; 20: 6638–6645.
Kim JY, Lee JY, Kim DG, Koo GB, Yu JW, Kim YS . TRADD is critical for resistance to TRAIL-induced cell death through NF-kappaB activation. FEBS Lett 2011; 585: 2144–2150.
Schneider F, Neugebauer J, Griese J, Liefold N, Kutz H, Briseno C et al. The viral oncoprotein LMP1 exploits TRADD for signaling by masking its apoptotic activity. PLoS Biol 2008; 6: e8.
Hatzivassiliou E, Miller WE, Raab-Traub N, Kieff E, Mosialos G . A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J Immunol 1998; 160: 1116–1121.
Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L . Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity 1997; 7: 821–830.
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 1997; 16: 5386–5397.
Bodmer JL, Meier P, Tschopp J, Schneider P . Cysteine 230 is essential for the structure and activity of the cytotoxic ligand TRAIL. J Biol Chem 2000; 275: 20632–20637.
Gasparian ME, Ostapchenko VG, Yagolovich AV, Tsygannik IN, Chernyak BV, Dolgikh DA et al. Overexpression and refolding of thioredoxin/TRAIL fusion from inclusion bodies and further purification of TRAIL after cleavage by enteropeptidase. Biotechnol Lett 2007; 29: 1567–1573.
Juo P, Woo MS, Kuo CJ, Signorelli P, Biemann HP, Hannun YA et al. FADD is required for multiple signaling events downstream of the receptor Fas. Cell Growth Differ 1999; 10: 797–804.
Hsu H, Shu HB, Pan MG, Goeddel DV . TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996; 84: 299–308.
Muppidi JR, Lobito AA, Ramaswamy M, Yang JK, Wang L, Wu H et al. Homotypic FADD interactions through a conserved RXDLL motif are required for death receptor-induced apoptosis. Cell Death Differ 2006; 13: 1641–1650.
Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ . A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 2006; 126: 995–1004.
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 2000; 1: 489–495.
Ting AT, Pimentel-Muinos FX, Seed B . RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J 1996; 15: 6189–6196.
Wachter T, Sprick M, Hausmann D, Kerstan A, McPherson K, Stassi G et al. cFLIPL inhibits tumor necrosis factor-related apoptosis-inducing ligand-mediated NF-kappaB activation at the death-inducing signaling complex in human keratinocytes. J Biol Chem 2004; 279: 52824–52834.
Kavuri SM, Geserick P, Berg D, Dimitrova DP, Feoktistova M, Siegmund D et al. Cellular FLICE-inhibitory protein (cFLIP) isoforms block CD95- and TRAIL death receptor-induced gene induction irrespective of processing of caspase-8 or cFLIP in the death-inducing signaling complex. J Biol Chem 2011; 286: 16631–16646.
Juo P, Kuo CJ, Yuan J, Blenis J . Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol 1998; 8: 1001–1008.
Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol 2010; 6: 352.
Tang W, Wang W, Zhang Y, Liu S, Liu Y, Zheng D . Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced chemokine release in both TRAIL-resistant and TRAIL-sensitive cells via nuclear factor kappa B. FEBS J 2009; 276: 581–593.
Varfolomeev E, Maecker H, Sharp D, Lawrence D, Renz M, Vucic D et al. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem 2005; 280: 40599–40608.
Cao X, Pobezinskaya YL, Morgan MJ, Liu Z-g . The role of TRADD in TRAIL-induced apoptosis and signaling. FASEB J 2011; 25: 1353–1358.
Kieser A . Pursuing different ‘TRADDes’: TRADD signaling induced by TNF-receptor 1 and the Epstein–Barr virus oncoprotein LMP1. Biol Chem 2008; 389: 1261–1271.
Wajant H, Scheurich P . TNFR1-induced activation of the classical NF-κB pathway. FEBS J 2011; 278: 862–876.
Choi C, Kutsch O, Park J, Zhou T, Seol DW, Benveniste EN . Tumor necrosis factor-related apoptosis-inducing ligand induces caspase-dependent interleukin-8 expression and apoptosis in human astroglioma cells. Mol Cell Biol 2002; 22: 724–736.
Harper N, Farrow SN, Kaptein A, Cohen GM, MacFarlane M . Modulation of tumor necrosis factor apoptosis-inducing ligand- induced NF-kappa B activation by inhibition of apical caspases. J Biol Chem 2001; 276: 34743–34752.
Wajant H, Haas E, Schwenzer R, Mühlenbeck F, Kreuz S, Schubert G et al. Inhibition of death receptor-mediated gene induction by a cycloheximide-sensitive factor occurs at the level of or upstream of fas-associated death domain protein (FADD). J Biol Chem 2000; 275: 24357–24366.
Kovalenko A, Kim JC, Kang TB, Rajput A, Bogdanov K, Dittrich-Breiholz O et al. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J Exp Med 2009; 206: 2161–2177.
Jin Z, El-Deiry WS . Distinct signaling pathways in TRAIL- versus tumor necrosis factor-induced apoptosis. Mol Cell Biol 2006; 26: 8136–8148.
Acknowledgements
We thank Ulrike Borgmeier for technical work, Katja Schneider for help in TRAIL production and Michela Carlet and Sebastian Tiedt for cell sorting. We also thank John Blenis for providing FADD- and caspase-8-deficient JURKAT cells and Elisabeth Kremmer for production of the HA antibody. RIP-deficient JURKAT cells were a kind gift from Brian Seed. This work was funded by Deutsche Forschungsgemeinschaft (SFB 684, TP-22), Deutsche Jose Carreras Leukämie Stiftung (R 10/26), Bettina Bräu Stiftung and Helmut Legerlotz Stiftung (all to IJ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellà
Supplementary Information accompanies the paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Grunert, M., Gottschalk, K., Kapahnke, J. et al. The adaptor protein FADD and the initiator caspase-8 mediate activation of NF-κB by TRAIL. Cell Death Dis 3, e414 (2012). https://doi.org/10.1038/cddis.2012.154
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.154
Keywords
This article is cited by
-
Cell death in head and neck cancer pathogenesis and treatment
Cell Death & Disease (2021)
-
Cell death in the gut epithelium and implications for chronic inflammation
Nature Reviews Gastroenterology & Hepatology (2020)
-
Extracellular vesicle-cargo miR-185-5p reflects type II alveolar cell death after oxidative stress
Cell Death Discovery (2020)
-
Stress-induced TRAILR2 expression overcomes TRAIL resistance in cancer cell spheroids
Cell Death & Differentiation (2020)
-
Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling
Cell Death & Disease (2019)