Abstract
microRNA-34a (miR-34a) and sirtuin 1 (SirT1) have been extensively studied in tumour biology and longevity/aging, but little is known about their functional roles in smooth muscle cell (SMC) differentiation from pluripotent stem cells. Using well-established SMC differentiation models, we have demonstrated that miR-34a has an important role in SMC differentiation from murine and human embryonic stem cells. Surprisingly, deacetylase sirtuin 1 (SirT1), one of the top predicted targets, was positively regulated by miR-34a during SMC differentiation. Mechanistically, we demonstrated that miR-34a promoted differentiating stem cells’ arrest at G0/G1 phase and observed a significantly decreased incorporation of miR-34a and SirT1 RNA into Ago2–RISC complex upon SMC differentiation. Importantly, we have identified SirT1 as a transcriptional activator in the regulation of SMC gene programme. Finally, our data showed that SirT1 modulated the enrichment of H3K9 tri-methylation around the SMC gene-promoter regions. Taken together, our data reveal a specific regulatory pathway that miR-34a positively regulates its target gene SirT1 in a cellular context-dependent and sequence-specific manner and suggest a functional role for this pathway in SMC differentiation from stem cells in vitro and in vivo.
Similar content being viewed by others
Main
Smooth muscle cell (SMC) differentiation is a critical process during cardiovascular system formation and development and has a significant role in cardiovascular system development and diseases. The understanding of the transcriptional regulatory circuitry of SMC differentiation is fundamental in stem cell therapy. However, the detailed molecular mechanisms of SMC differentiation have not been fully clarified. microRNAs (miRNAs) have been recently implicated in embryonic development, stem cell differentiation, cellular homeostasis and disease. Traditionally, mature miRNAs are thought to suppress gene expression by inducing mRNA cleavage1 or mRNA decay2 or by inhibiting mRNA translation;3 however, emerging evidence suggests that miRNAs can also upregulate target gene expression under specific circumstances.4, 5 Interestingly, miRNAs have been suggested to have an important role in cell/tissue specification due to the fact that many miRNAs are expressed in a tissue-specific manner.6 By disrupting Dicer7 or drosha8 expression in embryonic stem cells (ESCs), miRNAs are seen to have a role in stem cell self-renewal and differentiation. The significance of miRNAs in cardiovascular development has been revealed in a study of Dicer-deficient mice, which showed that the loss of miRNAs caused severe impairment of heart and blood vessel development.9 Furthermore, it has been shown that deletion of Dicer in vascular SMCs caused late embryonic lethality due to thinner vessel walls, impaired contractility and hemorrhage because of the decreased SMC proliferation and differentiation.10 However, the significance and exact role of individual miRNAs in SMC differentiation remain to be elucidated. In the present study, we demonstrate for the first time that microRNA-34a (miR-34a) has an important role in human and murine ESC differentiation towards SMC lineage. Unexpectedly, our data also reveal that miR-34a upregulates sirtuin 1 (SirT1), which is identified as a potential transcriptional activator of SMC genes.
Results
Important role of miR-34a in SMC differentiation from ESCs
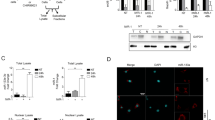
Murine ESCs were induced to differentiate towards SMCs as described previously (Supplementary Figure S1).11, 12, 13, 14 miRNA microarrays analysis was conducted to identify potential miRNA candidates for SMC differentiation, and the data revealed that miR-290 family members, the reported ESC-specific miRNA cluster,15 were dramatically downregulated upon differentiation (Supplementary Table S1). Conversely, muscle differentiation-related miRNAs (miR-143/145/133) were increased in our SMC differentiation model, while SMC proliferation-related miR-21 was undetectable at early stage but dramatically increased at late stage of SMC differentiation, indicating that some miRNAs may initiate SMC differentiation, whereas others may have important roles in the late stage of SMC differentiation. No significant changes were observed for other reported cell lineage-specific miRNAs (Supplementary Table S1). Interestingly, the microarray analysis showed that miR-34a, a reported cell cycle mediator,16 was upregulated to a greater magnitude than the best known SMC differentiation-related miRNAs miR-143 and -145 during SMC differentiation, which was further confirmed by RT-qPCR analysis (Figure 1a). The gain-of-function experiments showed that miR-34a overexpression induced the expression of SMC-specific genes (Figures 1b and c), while loss-of-function data revealed that knockdown of miR-34a inhibited the expression of SMC-specific markers at both RNA (Figure 1d) and protein levels (Figure 1e), suggesting an important role of miR-34a in SMC differentiation. Moreover, a microarray analysis was conducted to examine the gene expression profiling in differentiating SMCs affected by miR-34a overexpression. Expectedly, data summarised in Supplementary Table S2 showed that a panel of SMC-specific genes (highlighted in yellow) were upregulated by miR-34a overexpression. Importantly, several SMC-specific transcription factors, including serum response factor (SRF), myocardin (Myocd) and myocyte enhancer factor 2C (MEF2c), that were reported to be activated in our SMC differentiation system11, 17 were also significantly upregulated by miR-34a, alongside SirT1 (Supplementary Table S2). Furthermore, overexpression of miR-34a upregulated SRF, myocardin and MEF2c, whereas knockdown of miR-34a downregulated these genes (Supplementary Figure S2), suggesting that miR-34a works in concert with these SMC transcription factors during SMC differentiation.
Important role of miR-34a in SMC differentiation from ESCs. (a) miR-34a was upregulated during SMC differentiation from ESCs. Day-0 samples were undifferentiated ESCs and served as negative control. (b and c) miR-34a overexpression significantly increases SMC marker expression levels. ESCs were transfected with miR-34a precursor or miR precursor negative control and cultured in SMC DM for 48–72 h. miR-145 precursor in panel (c) was included as positive control. (d and e) miR-34a inhibition reduces SMC marker expression levels. Day-3 differentiating ESCs were transfected with miR-34a inhibitor or miR inhibitor negative control and cultured in SMC DM for 48 h. miR-145 inhibitor in panels (d and e) was included as positive control. The data presented here are representative or mean±S.E.M. of 3–6 independent experiments. *P<0.05
miR-34a is involved in SMC differentiation in vivo
To facilitate our in vivo cell implantation study, miR-34a overexpressing (pLL3.7-GFP-miR-34a) and control (pLL3.7-GFP) ESC lines were generated, and GFP-positive cells (Supplementary Figure S3A) were sorted out and maintained in ESC culture medium. Compared with control ESCs, the expression levels of miR-34a in miR-34a-overexpressing ESCs were slightly increased at day 0 but significantly upregulated upon differentiation (Supplementary Figure S3B), suggesting that miR-34a expression levels were successfully upregulated in these cells upon SMC differentiation. Consequently, more SMCs were differentiated from miR-34a-overexpressing ESCs (Supplementary Figure S3C), further confirming a promotive role of miR-34a in SMC differentiation. To explore the functional importance of miR-34a in SMC differentiation in vivo, pLL3.7-GFP and pLL3.7-GFP-miR-34a ESCs were subcutaneously injected into mice with 100 ng/ml of PDGF-BB to promote in vivo SMC differentiation as described in our previous study.11, 12 Data showed that more smooth muscle myosin heavy chain (SM-MHC)-positive cells were presented in the implants of miR-34a-overexpressing ESCs (Figures 2a and b) and that the majority of cells in the Matrigel implants were GFP-positive, indicating its exogenous origins (Figure 2a). Importantly, the expression levels of miR-34a, smooth muscle alpha-actin (SMαA), SM-MHC and SirT1 in the Matrigel implants of pLL3.7-GFP-miR-34a ESCs were significantly higher than that of control pLL3.7-GFP ESCs as demonstrated by immunofluorescence staining (Figure 2a), RT-qPCR (Figure 2c) and immunoblotting (Figure 2d) analyses, respectively, further confirming the efficiency of miR-34a overexpression and the importance of miR-34a in SMC differentiation in vivo.
miR-34a promotes SMC differentiation in vivo. Matrigel plugs implanted with ESCs without (pLL3.7-GFP) or with (pLL3.7-GFP-miR-34a) miR-34a overexpression were harvested, sectioned and subjected to immunofluorescence staining using antibodies against GFP and SM-MHC or SirT1 alone. (a) Representative images and (b) quantitative data of the percentage of SM-MHC-positive cells are presented here, respectively. Note: cells with green fluorescence signal indicate GFP-positive cells (implanted cells) within Matrigel plugs. The percentage of GFP-labelled SM-MHC-positive cells per field were examined by two well-trained independent investigators blinded to the treatments from four random high power fields ( × 200) in each section, three sections from each implant and four implants for each group, *P<0.05. (c and d) Gene expression levels within Matrigel implants. Total RNA and protein samples were extracted from partial Matrigel implants and subjected to RT-qPCR (c) and western blotting (d) analysis, respectively. The data presented here are representative or mean±S.E.M. of four Matrigel implants. *P<0.05
miR-34a also has an important role in SMC differentiation from human ESCs
To translate our finding from murine cell system to human, human ESCs were induced to differentiate towards SMCs using a similar differentiation protocol. Data showed that we have successfully differentiated human ESC into SMCs as demonstrated by RT-qPCR (Supplementary Figure S4A), western blotting (Supplementary Figure S4B), immunofluorescence staining (Supplementary Figure S4C) and flow cytometry (Supplementary Figure S4D) analyses. Along with SMC gene activation, miR-34a was significantly increased during SMC differentiation from human ESCs (Supplementary Figure S5A), implying that miR-34a may also have a role in human SMC differentiation. Importantly, data showed that miR-34a overexpression (Supplementary Figures S5B and C) significantly upregulated, while miR-34a inhibition (Supplementary Figures S5D and E) dramatically downregulated SMC gene expression levels in differentiating human ESCs, further demonstrating a critical role of miR-34a in human SMC differentiation.
SirT1 is positively regulated by miR-34a through the binding sites within 3′UTR
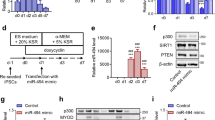
SirT1 was emerged as one of the top targets of miR-34a in several computational algorithmic databases, and two highly conserved binding sites for miR-34a have been identified within SirT1 3′UTR (Figure 3a). Unexpectedly, SirT1 gene expressions were significantly upregulated (Figure 3b) and displayed a positive correlation with the gene expression levels of miR-34a during SMC differentiation (Figure 1a), suggesting that miR-34a may positively regulate SirT1. Such notion has been firmly supported by the findings that SirT1 gene and protein levels were significantly upregulated or downregulated by overexpression or inhibition of miR-34a in the differentiating ESCs (Figures 3c and d), respectively. Importantly, our miRNA reporter assay showed that the activity of luciferase from construct harbouring the wild-type, but not the binding site 2 mutant, of SirT1 3′UTR was significantly upregulated by miR-34a overexpression, implying that the binding site 2 is responsible for SirT1 3′UTR reporter activity upregulation mediated by miR-34a (Figure 3e). Altogether, above data has firmly confirmed that SirT1 is a true mRNA target of miR-34a, which is positively regulated by miR-34a during SMC differentiation.
Target gene, SirT1, is positively regulated by miR-34a through the binding sites within 3′ untranslated region (UTR). (a) The potential binding sites of miR-34a within SirT1 3′UTR as predicted by Targetscan are depicted in this illustration. (b) SirT1 was upregulated during SMC differentiation from ESCs. (c and d) Modulations of miR-34a expression levels positively regulate SirT1. (e) miR-34a-binding site 2 is required for miR-34a-mediated SirT1 gene activation. miR-34a precursor or negative control and wild-type SirT1 3′UTR reporter (pmiR-Luc-SirT1-WT) or three mutants (bindings site 1 (BS1mu), 2 (BS2mu) and combinational mutations (BS1/2mu)) were co-transfected into days 2–3 differentiating ESCs, and luciferase activity assay were measured at 48 h posttransfection. The data presented here are representative or mean±S.E.M. of 3–4 independent experiments. *P<0.05 (treatment versus day 0 or control), #P<0.05 in panel (e) (binding site mutants versus wild type)
miR-34a differently regulates SirT1 gene expression in ESCs and terminally differentiated SMCs
To investigate whether the observed phenomena that miR-34a directly upregulated SirT1 expression during SMC differentiation is cellular context specific, we conducted miR-34a overexpression or inhibition experiments and luciferase activity assays in naive stem cells (ESCs) and terminally differentiated SMCs (dSMCs; we have previously reported that day-8 dSMCs are terminally dSMCs17), respectively. Data showed that SirT1 3′UTR reporter activity was significantly downregulated by miR-34a overexpression but upregulated by miR-34a inhibition in ESCs (Figure 4a). As expected, SirT1 gene expression was significantly increased by overexpression of miR-34a, whereas decreased by miR-34a knockdown in terminally dSMCs (Figure 4b), suggesting that miR-34a regulates SirT1 gene expression is cellular context dependent. As the miR-34a-binding sites are required for miR-34a-mediated SirT1 gene regulation (Figure 3e), we further wondered whether the miR-34a-binding site is sufficient for miR-34a-mediated SirT1 gene expression. To this aim, we constructed a luciferase reporter with an artificial miR-34a-binding site without surrounding RNA sequence and structure of SirT1 3′UTR, designated as pmiR-Luc-miR-34a-binding site, and performed luciferase activity assay. We found that miR-34a slightly upregulated this reporter activity, but could not reach significance (Figure 4c), indicating that the surrounding RNA sequence and/or structure of SirT1 3′UTR is also required for miR-34a-mediated SirT1 gene upregulation.
miR-34a differently regulates SirT1 gene expression in ESCs and terminally differentiated SMCs. (a) miR-34a negatively regulates SirT1 expression in naive stem cell stage. Undifferentiated ESCs were co-transfected with control miRNAs, miR-34a precursor, or miR-34a inhibitor and pmiR-Luc-SirT1, respectively. Luciferase activity assay were measured at 48 h posttransfection. (b) miR-34a positively regulates SirT1 expression in terminally differentiated stage. Day-8 differentiated SMCs were co-transfected with control miRNAs, miR-34a precursor, or miR-34a inhibitor and pmiR-Luc-SirT1, respectively. Luciferase activity assay were measured at 48 h posttransfection. (c) Surrounding structure(s) or sequences of SirT1 3′ untranslated region (UTR) is required for miR-34a-mediated SirT1 gene upregulation. (d) Ago2 protein levels and IP efficiency. The expression levels of Ago2 (input) and RNA immunoprecipitation (RNA-IP) efficiencies (Ago2-IP and IgG-IP) in undifferentiated ESCs (ESCs) and differentiated SMCs (dSMCs) were examined using Ago2 antibody. (e) Less miR-34a and SirT1 mRNA were incorporated into Ago2–RISC complex upon differentiation. Undifferentiated ESCs and differentiated SMCs were harvested and subjected to RNA-IP assay. RNA-IP assays were performed using antibodies against Ago2 or normal rabbit IgG, respectively, as described in online Supplementary Data. RT-qPCR amplification of the SirT1 coding region was included as additional control for specific SirT1 3′UTR enrichment. Dashed line in panel (e) indicates the enrichment level for IgG control, which was set as 1.0. The data presented here are representative or mean±S.E.M. of 3–4 independent experiments. *P<0.05 (versus control)
We have previously reported that miR-200c mediates endothelial cell differentiation by targeting zinc finger E-box-binding homeobox 1 (ZEB1),18 which contains no miR-34a-binding site within 3′UTR. Expectedly, we observed no effects of miR-34a overexpression on ZEB1 gene reporter activity (Supplementary Figure S6A), confirming that miR-34a specifically upregulates SirT1 gene expression during SMC differentiation. To further distinguish whether our observation that upregulation of SirT1 by miR-34a during SMC differentiation is a universal phenomenon or a specific event, we performed luciferase assays with the non-miR-34a reporter (pmiR-Luc-ZEB1) in conjunction with miR-200c overexpression in ESCs and terminally dSMCs, respectively. We found that miR-200c negatively regulated ZEB1 gene reporter activity in both cells (Supplementary Figures S6B and C), which is consistent with our previous observation in endothelial cell differentiation. Moreover, it is well known that 12-O-tetra-decanoylphorbol-13-acetate (TPA) can induce monocyte differentiation through regulating cell cycle progression. We then applied this model to investigate whether upregulation of SirT1 by miR-34a may (or may not) be specific to differentiating stem cells. Although miR-34a expression levels were clearly upregulated by TPA during monocyte differentiation, no significant changes in terms of SirT1 expression levels were observed during the same process (Supplementary Figure S6D), implying no significant involvement of miR-34a in regulation of SirT1 gene expression in this monocyte differentiation model. Taken together, above data clearly suggest that upregulation of SirT1 by miR-34a during SMC differentiation is a specific event, which is likely dependent on specific cellular context, miRNA-binding sites and surrounding sequence.
Less miR-34a and SirT1 mRNA were incorporated into argonaute 2 (Ago2)–RISC (RNA-induced silencing complex) complex upon differentiation
To further investigate the mechanism by which miR-34a upregulates SirT1 gene expression upon SMC differentiation, we conducted the RNA-immunoprecipitation assays with Ago2 antibody in both ESCs and dSMCs. Expectedly, no significant difference in terms of Ago2 protein levels between ESCs and dSMCs was observed (input samples, Figure 4d), and the immunoprecipitation efficiencies in both cells are comparable (Ago2-IP and IgG-IP samples, Figure 4d). Moreover, compared with IgG control Ago2 antibody could specifically pull down much more miR-34a and SirT1 RNA (Figure 4f). Importantly, the enrichment levels of miR-34a and SirT1 RNA (spanning around miR-34a-binding sites 1 and 2) in ESCs were much higher than that of dSMCs (Figure 4e), indicating that much less miR-34a and SirT1 3′UTR were loaded into Ago2–RISC complex upon SMC differentiation.
miR-34a promotes cell cycle arrest at G0/1 in differentiating ESCs
Cell cycle analyses revealed that differentiating cells were gradually arrested at G0/1 phase during SMC differentiation (Supplementary Figure S7A), which was significantly reinforced by miR-34a overexpression (Supplementary Figure S7B). Moreover, data showed that miR-34a has no significant role in cell apoptosis and death (Supplementary Figure S7C), while much less cells were entering S-phase upon miR-34a overexpression (Supplementary Figure S7D). To further confirm whether miR-34a-upregulated SirT1 gene expression is specific to G0/G1 cell cycle stage, differentiating SMCs were arrested at G0/G1 phase by cell cycle arrest inducer aphidicolin,4 and luciferase activity assays were conducted. As expected, incubation of differentiating SMCs with 2 μg/ml aphidicolin for 24–28 h significantly increased cells arrest at G0/G1 stage, which was further increased in the presence of miR-34a overexpression (Supplementary Figure S7E). Consequently, SirT1 3′UTR reporter activity was significantly upregulated by aphidicolin and miR-34a overexpression in a similar manner (Supplementary Figure S7F).
SirT1 activation is required for miR-34a-mediated SMC gene expression levels
Data from SirT1 overexpression experiments with pcDNA3.1-SirT1 plasmid (kindly provided by Dr. Hang Shi19) showed that SirT1 significantly upregulated SMC marker expression levels (Figures 5a and b), suggesting that SirT1 overexpression can recapitulate the effects of miR-34a during SMC differentiation. We further hypothesised that SirT1 activity is required for miR-34a-mediated SMC differentiation. To test this, pLL3.7-GFP or pLL3.7-GFP-miR-34a ESCs were induced to differentiate into SMCs in the absence or presence of SirT1-specific inhibitor. Data shown in Figure 5c revealed that while SirT1 inhibition (second columns) or miR-34a overexpression (third columns) alone in the differentiating ESCs significantly downregulated or upregulated various SMC-specific gene expression, respectively, inhibition of SirT1 almost abolished SMC-specific gene upregulations induced by miR-34a overexpression (fourth columns). Such observation was further confirmed in SirT1 knockdown experiments by using SirT1-specific siRNA (Figure 5d), suggesting that miR-34a regulates SMC gene expression during SMC differentiation from ESCs through activation of SirT1. It has been reported that SirT1 agonist resveratrol promotes osteogenic differentiation of mesenchymal stem cells.20, 21 Consistently, our data revealed that resveratrol also significantly increased SMC-specific gene expression (Figure 5e), further supporting a role of SirT1 activation in SMC differentiation.
SirT1 activation is required for miR-34a-mediated SMC gene expression levels. (a and b) Overexpression of SirT1-upregulated SMC gene expression levels. Days 2–3 differentiating ESCs were transfected with control (pcDNA3.1) or SirT1 overexpression plasmid (pcDNA3.1-SirT1) and cultured in SMC DM for another 48 or 72 h. The data presented here are representative or mean±S.E.M. of three independent experiments, *P<0.05. (c) SirT1 inhibition abolished SMC gene expression increased by miR-34a overexpression. Control (pLL3.7-GFP) and miR-34a overexpressing (pLL3.7-GFP-miR-34a) ESCs were induced to differentiate into SMCs for 4 days, and SirT1-specific inhibitor III (500 nM, EMD Millipore, 566322) or control vehicle were added into the culture medium for 6 or 12 h before harvest. (d) SirT1 knockdown abrogated the effects of miR-34a overexpression on SMC gene expression. Days 2–3 differentiating ESCs were co-transfected with miR-34a precursor, SirT1 siRNAs or respective control (miRNA precursor negative control and siRNA control) as indicated and cultured in SMC DM for another 48 h. (e) SirT-1 agonist resveratrol promotes SMC gene expression. The data presented here are mean±S.E.M. of 3–4 independent experiments. *P<0.05 (versus related controls); #P<0.05 (4th bars versus third bars)
SRF-binding site is required for SirT1-mediated SMC gene expression
Luciferase activity assays with pGL3-Luc-SMαA22 and pGL3-Luc-SM22α22 showed that the overexpression of SirT1 significantly increased SMαA and SM22α gene-promoter activities (Figure 6a) and that SRF-binding element (CArG) mutation11 in pGL3-Luc-SMαA and pGL3-Luc-SM22α almost abolished their transcriptional activity in response to SirT1 overexpression (Figure 6a), suggesting that SirT1 regulates SMC differentiation gene expression through SRF-binding site(s). Moreover, chromatin immunoprecipitation (ChIP) assays revealed that SirT1 directly bound to the region spanning around SRF-binding element of SMαA and SM22α gene promoters and that such binding was dramatically enhanced by SirT1 overexpression (Figure 6b). Finally, ChIP assay data also showed that the binding capacity of SRF to SMαA and SM22α gene promoters was significantly enhanced by SirT1 overexpression (Figure 6c). Taken together, above findings demonstrate that SirT1 regulates SMC-specific gene expression levels during SMC differentiation through direct binding to the promoter region of SMαA and SM22α genes and increasing the binding ability of SRF to SMC-specific gene promoters.
SRF-binding site is required for SirT1-mediated SMC gene expression. (a) SRF-binding site mutation abolished SMC differentiation gene-promoter activity induced by SirT1 overexpression. (b) SirT1 binds directly to the promoter regions of SMC differentiation genes. (c) Upregulated SirT1 increases SRF binding to the promoter regions of SMC differentiation genes. ChIP assays were performed using antibodies against SirT1, SRF, or normal rabbit IgG, respectively, as described in online Supplementary Data. PCR amplifications of the non-CArG regions were included as additional control for specific promoter DNA enrichment. The data presented here are mean±S.E.M. of four independent experiments. *P<0.05 (versus control)
SMC transcription factors are transcriptionally regulated by SirT1
SRF, MEF2c and myocardin are well-known transcription factors for regulating SMC gene expression. Data shown in Figure 7a revealed that gene expression levels of all three factors were significantly upregulated by SirT1, suggesting SirT1 may have a direct role in regulation of these transcription factors during SMC differentiation. Indeed, luciferase activity assays with SRF, MEF2c and Myocd gene reporters11 showed that the SirT1 overexpression significantly increased SRF, MEF2c or Myocd gene-promoter activities (Figure 7b), indicating that SirT1 upregulates transcriptional activity of these three genes. Importantly, data shown in Figure 7c revealed that SirT1 directly bound to the promoter regions between −1393 and −1274 of SRF gene, −1335 and −1263 of MEF2c gene or −708 and −620 of Myocd gene, respectively, and that such binding activity was significantly enhanced by SirT1 overexpression. Taken together, these findings strongly imply that SirT1 transcriptionally regulates SMC transcription factor gene expression during SMC differentiation.
SMC transcription factor gene expression levels were positively and transcriptionally regulated by SirT1. (a) mRNA levels of SRF, MEF2c and myocd were significantly upregulated by SirT1 overexpression. Total RNAs were harvested as described in Figure 5a. (b) SirT1 regulated the promoter activities of SMC transcription factor genes. Days 2–3 differentiating ESCs were transfected with luciferase reporter plasmids pGL3-SRF-Luc, pGL3-MEF2c-Luc, or pGL3-Myocd-Luc (0.15 μg/2.5 × 104 cells) together with pcDNA3.1-SirT1 or pcDNA3.1 (0.2 μg/2.5 × 104 cells). pShuttle-LacZ (0.2 μg/2.5 × 104 cells) was included as control. Luciferase and β-galactosidase activity assays were detected 48 h after transfection. The data presented here are mean±S.E.M. of 3–6 independent experiments. *P<0.05 (versus control). (c) SirT1 binds directly to the promoter regions of SRF, MEF2c and myocardin genes. ChIP assays were performed using antibodies against SirT1 or normal rabbit IgG, respectively, as described in online Supplementary Data. PCR amplifications of the adjacent regions were included as additional control for specific promoter DNA enrichment. The data presented here are mean±S.E.M. of four independent experiments. *P<0.05 (versus control)
SirT1 promotes SMC gene expression through inhibiting H3K9 methylation around SMC gene promoters
Methylation of lysines H3K9 and H3K27 is closely associated with transcriptional repression.23 Data obtained from SirT1 overexpression experiments showed that H3K9me3 protein expression levels were not regulated by SirT1 overexpression (Supplementary Figure S8A); however, their enrichments on SMαA and SM22α gene promoters were significantly inhibited by overexpression of SirT1 (Supplementary Figure S8B), suggesting that SirT1 regulates SMC-specific gene activation, at least partially through inhibiting H3K9 tri-methylation around SMC-specific gene promoters.
Discussion
miR-34 family comprises three processed miRNAs: miR-34a, miR-34b, and miR-34c.24 Members of the miR-34 family have been suggested as critical regulators in regulation of cancer cell apoptosis and cell cycle arrest.16, 25, 26 It has been recently reported that suppression of somatic cell reprogramming into pluripotent cells by miR-34a was due to their repression of pluripotency genes,27 implying that miR-34a could have an important role in stem cell differentiation. Indeed, recent studies show that miR-34a has a role in neural28 and megakaryocytic differentiation29 and is a critical cell-fate determinant in early-stage dividing colon cancer stem cells.30 In the present study, we provide compelling evidence to support a critical role of miR-34a in SMC differentiation.
It has been reported that miR-34a regulates cell cycle progress and apoptosis in cancer cells by repressing SirT1 and forming a double-positive feedback loop to regulate p53 activity.31, 32 Surprisingly, we have provided solid evidence to suggest that, rather than translational repression, miR-34a regulates SirT1 in a translational activation manner. However, it is noteworthy to point out that the upregulation of SirT1 expression by miR-34a during SMC differentiation is representing a specific event between miR-34a and SirT1 in our SMC differentiation model, which is likely dependent on specific cellular context, miRNA-binding sites and surrounding sequence. Such notion has been supported by following evidence. First, SirT1 gene and protein expression levels were positively regulated by miR-34a expression levels. Second, overexpression of miR-34a significantly upregulates SirT1 3′UTR activity, but such upregulation was completely abolished when the miR-34a-binging sites were mutated. Third, the luciferase activity of non-miR-34a target gene reporters was not regulated by miR-34a. Fourth, SirT1 gene expression level is positively associated with miR-34a expression level during in vivo SMC differentiation. Finally, we observed a negative regulatory relationship between a non-miR-34a reporter and its corresponding miRNA during SMC differentiation. The unexpected but novel finding that miR-34a upregulates SirT1 could be explained by the following observations: (1) Differentiating ESCs undergo cell cycle arrest at G0/1 and miR-34a promotes cell cycle arrest at G0/1 phase during SMC differentiation. As mentioned early, the miRNA-target gene regulatory machinery will switch from translation repression to activation when the cells have been arrested at G0/1 phase4 or locked in quiescent status.5 (2) Another suggested criteria by the study4 for miRNAs upregulate their target genes is the AU-rich elements existing within 3′UTR. By using online search tool (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi), we have found that SirT1 3′UTR is an A/U-rich sequence (67%) and identified nearly 11 AU-rich elements (ARE) sites within 3′UTR of SirT1. Most importantly, we have also found that the miR-34a-binding site 2 is located closely with 8 of the 11 ARE sites within SirT1 3′UTR (Supplementary Figure S9). (3) miR-34a negatively regulates SirT1 gene expression in ESCs, whereas positively regulates SirT1 gene expression in the terminally dSMCs. (4) A synergetic effect between aphidicolin and miR-34a overexpression on SirT1 gene regulation was observed. (5) Removal of surrounding RNA sequence and structure of SirT1 3′UTR almost abolished the upregulative effects of miR-34a on SirT1 3′UTR reporter activity. All these observations have provided compelling evidence to suggest that upregulation of SirT1 by miR-34a during SMC differentiation is a specific event between miR-34a and SirT1 gene during SMC differentiation and that both miR-34a-binging sites and surrounding RNA sequence/structures within 3′UTR of SirT1 gene are required for such an event.
Well-documented evidence suggests that RNA-binding proteins (RBPs) have a crucial role in modulating miRNA function. Kundu et al.33 have nicely demonstrated that RNA-binding protein HuR attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Moreover, our previous studies have suggested an important role for another two RBPs, hnRNPA111 and hnRNPA2B1,34 in SMC differentiation. These evidence together with our observation that much less miR-34a and SirT1 3′UTR were loaded into Ago2–RISC complex upon SMC differentiation prompts us to speculate that miR-34a upregulates SirT1 during SMC differentiation through a similar mechanism as reported by Kundu et al.33 In other words, the upregulation of SirT1 by miR-34a may well be a result of RBPs, such as HuR, hnRNPA1 and hnRNPA2B1, interacting with miR-34a-binding sites and/or other elements within the 3′UTR of SirT1 to disengage it from Ago2–RISC complex during differentiation. Although it warrants further study, investigations into confirming such a possibility is outside the current remit of this study.
Another novel mechanistic finding in the present study is that we provide clear evidence to support that SirT1 regulates SMC differentiation gene expression through a transcriptional mechanism. SirT1 has been reported to be involved in a variety of cellular functions.35 Recent data also suggests that SirT1 is a critical mediator in regulation of various development genes during stem cell differentiation36 and has an important role in various cellular differentiations, including endothelial progenitor cells,37 hematopoietic cells38 and osteoblasts.39 Traditionally, SirT1 has mainly been linked to gene repression;40 however, accumulating evidence also strongly suggests that SirT1 can act both positively and negatively to control gene expression by recruiting a different set of coactivators and corepressors41 or via a transcriptional mechanism.42 In consistent with these findings, in the current study we have demonstrated for the first time that SirT1 is a transcription activator in regulation of SMC-specific gene expression during SMC differentiation.
Taken together, we have successfully uncovered a novel role of miR-34a in SMC differentiation and provided compelling evidence to support our unexpected finding that miR-34a positively regulates its target gene SirT1 during SMC differentiation. Moreover, we have revealed that SirT1 regulates SMC gene expression through a transcriptional mechanism as well as an epigenetic signal pathway. Furthermore, our data also suggests that the miRNA-target gene regulatory machinery should be carefully considered under certain circumstances. These findings significantly increase our understanding of the molecular mechanisms in SMC differentiation and will benefit future applications in regenerative medicine.
Materials and Methods
ESC culture and SMC differentiation
Detailed protocols for mouse ESCs (mESCs) (ES-D3 cell line, CRL-1934; ATCC, Manassas, VA, USA) culture and SMC differentiation were described in our previous studies.11, 12, 13, 14, 17, 22, 34, 43, 44 The Shef-1, 2, 3, 6, and 7 human ESC lines were obtained from the United Kingdom Stem Cell Bank (UKSCB, Hertfordshire, UK) and maintained in our Laboratory as described in our previous study.18 Briefly, undifferentiated ESCs were dissociated into single cells and seeded onto collagen I/IV (5 μg/ml)-coated flasks or plates in differentiation medium (DM, MEM alpha medium (Gibco, Carlsbad, CA, USA) supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin) for 0–9 days prior to further treatment. The medium was refreshed every other day. In our preliminary studies, we observed that there were no significant differences in terms of SMC differentiation capacity among these cell lines, but the Shef-3 cell line was the best to grow and scale-up. Therefore, Shef-3 cell line was routinely maintained in our laboratory and used in the current study.
miRNA and plasmids transfection
Either miRNAs inhibitors or precursors and miRNA-negative controls (30 nM) were transfected into differentiating ESCs using siPORT NeoFX transfection agent (Ambion, Foster City, CA, USA) according to the manufacturer’s instructions. Transfected cells were plated into flasks or plates coated with 5 μg/ml of collagen and cultured for 48–72 h in the SMC DM to allow SMC differentiation. All miRNA inhibitors or precursors and respective negative controls were purchased from Ambion. Control (pcDNA3.1) and SirT1 overexpression (pcDNA3.1-SirT1,19 a kind gift from Dr. Hang Shi, Wake Forest University School of Medicine, NC, USA) plasmids were transfected into differentiating ESCs using Fugene-6 (Roche, Basel, Switzerland) according to the manufacturer’s instructions.
SirT 3′UTR clone and miR-34a-binding site mutation
Reporter vector harbouring sequences of the murine SirT1 was created using cDNA from differentiating ESCs. The 3′-flanking untranslation region (2306bp/3636bp) of murine SirT1 gene (NM_019812.2) was amplified by PCR with primers shown in Supplementary Table S3 and cloned into the Mlu I and Sac I sites of the pmiR-reporter-basic vector (Ambion), designated as pmiR-Luc-SirT1-WT. miR-34a-binding sites 1, 2 mutation alone or combination were introduced into pmiR-Luc-SirT1 by using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. These were designated as pmiR-Luc-SirT1-BS1mu, pmiR-Luc-SirT1-BS2mu, and pmiR-Luc-SirT1-BS1/2mu mutants, respectively. An artificial miR-34a-binding site was introduced into pmiR-Luc by using the PCR-based QuikChange site-directed mutagenesis, designated as pmiR-Luc-miR-34a-binding site. All vectors were verified by DNA sequencing.
Statistical analysis
Data were expressed as mean±S.E.M. and analyzed using a two-tailed Student’s t-test for two-group comparision or one-way ANOVA followed by Tukey’s HSD multiple comparison post-hoc test for comparing different groups. A value of P<0.05 was considered as statistically significant.
A detailed description on the expanded Materials and Methods is provided in the online Supporting Information.
Abbreviations
- ESC:
-
embryonic stem cells
- SMC:
-
smooth muscle cell
- miRNA:
-
microRNA
- miR-34a:
-
microRNA-34a
- SmαA:
-
smooth muscle alpha-actin
- SM-MHC:
-
smooth muscle myosin heavy chain
- ZEB1:
-
zinc finger E-box-binding homeobox 1
- SirT1:
-
sirtuin 1
- SRF:
-
serum response factor
- Myocd:
-
myocardin
- MEF2c:
-
myocyte enhancer factor 2C
- TPA:
-
12-O-tetra-decanoylphorbol-13-acetate
- Ago2:
-
argonaute 2
- CHIP:
-
chromatin immunoprecipitation
- RISC:
-
RNA-induced silencing complex
References
Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005; 122: 553–563.
Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312: 75–79.
Filipowicz W, Bhattacharyya SN, Sonenberg N . Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114.
Vasudevan S, Tong Y, Steitz JA . Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318: 1931–1934.
Vasudevan S, Tong Y, Steitz JA . Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 2008; 7: 1545–1549.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129: 1401–1414.
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ . Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 2005; 102: 12135–12140.
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R . DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007; 39: 380–385.
Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G . Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 2005; 280: 9330–9335.
Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC . MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol 2010; 30: 1118–1126.
Huang Y, Lin L, Yu X, Wen G, Pu X, Zhao H et al. Functional involvements of heterogeneous nuclear ribonucleoprotein A1 in smooth muscle differentiation from stem cells in vitro and in vivo. Stem Cells 2013; 31: 906–917.
Xiao Q, Pepe AE, Wang G, Luo Z, Zhang L, Zeng L et al. Nrf3-Pla2g7 interaction plays an essential role in smooth muscle differentiation from stem cells. Arterioscler Thromb Vasc Biol 2012; 32: 730–744.
Xiao Q, Wang G, Yin X, Luo Z, Margariti A, Zeng L et al. Chromobox protein homolog 3 is essential for stem cell differentiation to smooth muscles in vitro and in embryonic arteriogenesis. Arterioscler Thromb Vasc Biol 2011; 31: 1842–1852.
Pepe AE, Xiao Q, Zampetaki A, Zhang Z, Kobayashi A, Hu Y et al. Crucial role of nrf3 in smooth muscle cell differentiation from stem cells. Circ Res 2010; 106: 870–879.
Houbaviy HB, Murray MF, Sharp PA . Embryonic stem cell-specific MicroRNAs. Dev Cell 2003; 5: 351–358.
He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Xiao Q, Luo Z, Pepe AE, Margariti A, Zeng L, Xu Q . Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am J Physiol Cell Physiol 2009; 296: C711–C723.
Luo Z, Wen G, Wang G, Pu X, Ye S, Xu Q et al. MicroRNA-200C and -150 play an important role in endothelial cell differentiation and vasculogenesis by targeting transcription repressor ZEB1. Stem Cells 2013; 31: 1749–1762.
Yang Z, Kahn BB, Shi H, Xue BZ . Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem 2010; 285: 19051–19059.
Shakibaei M, Shayan P, Busch F, Aldinger C, Buhrmann C, Lueders C et al. Resveratrol mediated modulation of Sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: potential role of Runx2 deacetylation. PLoS One 2012; 7: e35712.
Tseng PC, Hou SM, Chen RJ, Peng HW, Hsieh CF, Kuo ML et al. Resveratrol promotes osteogenesis of human mesenchymal stem cells by upregulating RUNX2 gene expression via the SIRT1/FOXO3A axis. J Bone Miner Res 2011; 26: 2552–2563.
Margariti A, Xiao Q, Zampetaki A, Zhang Z, Li H, Martin D et al. Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J Cell Sci 2009; 122: 460–470.
Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ . Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics 2009; 10: 143.
Hermeking H . The miR-34 family in cancer and apoptosis. Cell Death Differ 2010; 17: 193–199.
Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007; 26: 745–752.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007; 26: 731–743.
Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011; 13: 1353–1360.
Aranha MM, Santos DM, Sola S, Steer CJ, Rodrigues CM . miR-34a regulates mouse neural stem cell differentiation. PLoS One 2011; 6: e21396.
Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood 2009; 114: 2181–2192.
Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 2013; 12: 602–615.
Yamakuchi M, Ferlito M, Lowenstein CJ . miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 2008; 105: 13421–13426.
Yamakuchi M, MiR-34 Lowenstein CJ . SIRT1 and p53: the feedback loop. Cell Cycle 2009; 8: 712–715.
Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W . HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 2012; 40: 5088–5100.
Wang G, Xiao Q, Luo Z, Ye S, Xu Q . Functional impact of heterogeneous nuclear ribonucleoprotein A2/B1 in smooth muscle differentiation from stem cells and embryonic arteriogenesis. J Biol Chem 2012; 287: 2896–2906.
Yang Yang WD, Yue Li, Zhenxiao Jin, Juanjuan Yan, Shiqiang Yu, Dinghua Yi . Novel role of silent information regulator 1 in myocardial ischemia. Circulation 2013; 128: 2232–2240.
Calvanese V, Lara E, Suarez-Alvarez B, Abu Dawud R, Vazquez-Chantada M, Martinez-Chantar ML et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci USA 2010; 107: 13736–13741.
Cheng BB, Yan ZQ, Yao QP, Shen BR, Wang JY, Gao LZ et al. Association of SIRT1 expression with shear stress induced endothelial progenitor cell differentiation. J Cell Biochem 2012; 113: 3663–3671.
Ou X, Chae HD, Wang RH, Shelley WC, Cooper S, Taylor T et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood 2011; 117: 440–450.
Srivastava S, Bedi U, Roy P . Synergistic actions of insulin-sensitive and Sirt1-mediated pathways in the differentiation of mouse embryonic stem cells to osteoblast. Mol Cell Endocrinol 2012; 361: 153–164.
Tissenbaum HA, Guarente L . Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 2001; 410: 227–230.
Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P . Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005; 434: 113–118.
Chang HC, Guarente L . SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013; 153: 1448–1460.
Zhang L, Jin M, Margariti A, Wang G, Luo Z, Zampetaki A et al. Sp1-dependent activation of HDAC7 is required for platelet-derived growth factor-BB-induced smooth muscle cell differentiation from stem cells. J Biol Chem 2010; 285: 38463–38472.
Xiao Q, Zeng L, Zhang Z, Hu Y, Xu Q . Stem cell-derived Sca-1+ progenitors differentiate into smooth muscle cells, which is mediated by collagen IV-integrin alpha1/beta1/alphav and PDGF receptor pathways. Am J Physiol Cell Physiol 2007; 292: C342–C352.
Acknowledgements
We are grateful to the supports from the British Heart Foundation (FS/09/044/28007, PG/11/40/28891, and PG/13/45/30326), National Natural Science Foundation of China (91339102, 81270001, and 81270180), and Zhejiang Provincial Natural Science Foundation (LR14H020001). This work forms part of the research themes contributing to the translational research portfolio of Barts and the London Cardiovascular Biomedical Research Unit, which is supported and funded by the National Institute of Health Research.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, X., Zhang, L., Wen, G. et al. Upregulated sirtuin 1 by miRNA-34a is required for smooth muscle cell differentiation from pluripotent stem cells. Cell Death Differ 22, 1170–1180 (2015). https://doi.org/10.1038/cdd.2014.206
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.206
This article is cited by
-
MicroRNA-146a Promotes Embryonic Stem Cell Differentiation towards Vascular Smooth Muscle Cells through Regulation of Kruppel-like Factor 4
Current Medical Science (2023)
-
MiR-140-3p inhibits the cell viability and promotes apoptosis of synovial fibroblasts in rheumatoid arthritis through targeting sirtuin 3
Journal of Orthopaedic Surgery and Research (2021)
-
miR-34c inhibits PDGF-BB-induced HAVSMCs phenotypic transformation and proliferation via PDGFR-β/SIRT1 pathway
Molecular Biology Reports (2021)
-
miR-194-5p negatively regulates the proliferation and differentiation of rabbit skeletal muscle satellite cells
Molecular and Cellular Biochemistry (2021)
-
miR-214-3p-Sufu-GLI1 is a novel regulatory axis controlling inflammatory smooth muscle cell differentiation from stem cells and neointimal hyperplasia
Stem Cell Research & Therapy (2020)