Abstract
Aim:
To explore a novel function of a mutant of the hepatitis B virus X protein (HBxΔ127) in the promotion of hepatoma cell migration.
Methods:
The effect of HBxΔ127 and wild type HBx on the migration ability of hepatoblastoma HepG2 cells were examined using wound healing assays in stable transfection systems. The full-length osteopontin(OPN) promoter sequence was cloned into the pGL3-Basic plasmid. The promoter activities of OPN in stably HBxΔ127-transfected hepatoblastoma HepG2 (HepG2-XΔ127) and hepatocellular carcinoma H7402 (H7402-XΔ127) cells were determined using luciferase reporter gene assays. The mRNA expression levels of OPN were detected by RT-PCR. And the effect of MK886, a specific inhibitor of 5-lipoxygenase (5-LOX), on OPN promoter activity and mRNA expression in HepG2-XΔ127 and H7402-XΔ127 cells were examined using luciferase reporter gene assays and RT-PCR, respectively. Finally, the migration ability of HepG2-XΔ127 was observed after treatment with siRNA targeting OPN mRNA and HBx mRNA using wound healing assays.
Results:
HepG2-XΔ127 cells exhibited a greater capacity for wound repair compared to HepG2-X cells. The promoter activity and mRNA expression levels of OPN were also increased in HepG2-XΔ127 and H7402-XΔ127 cells. Moreover, MK886 abolished the HBxΔ127-mediated upregulation of OPN. Wound healing assays demonstrated that the migration ability of HepG2-XΔ127 cells can be suppressed by treatment with siRNA targeting OPN mRNA and siRNA targeting HBx mRNA.
Conclusion:
HBxΔ127 strongly promotes hepatoma cell migration via activation of OPN involving 5-LOX.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in China. Infection by the hepatitis B virus (HBV) is a major risk factor for development of HCC1. The HBV genome includes four partially overlapping open reading frames termed the preC/C, P, preS/S, and X genes. Hepatitis B virus X protein (HBx) is encoded by the X gene, which contributes to the development of HCC2. HBx is an important regulator that has been shown to have multiple biological functions, including transcriptional activation of a variety of viral and cellular promoters, interaction with other proteins, mediation of cell proliferation, and activation of apoptosis3, 4, 5. Several studies have reported that HBx promotes invasion and metastasis in HBV-associated HCC by inducing matrix metalloproteinase (MMP) activation and, eventually, inducing the destruction of the extracellular matrix6, 7, 8. Moreover, HBx facilitates integrin-mediated cell migration and mediates the adhesion/deadhesion balance of cells at the primary tumor site9. HBx enhances CD44-mediated HA-interaction efficiency and modifies the migratory behavior of transformed heptocytes10. It has been reported that mutations in the HBx gene, especially the COOH-terminal deletion of HBx, are frequent events associated with the development of HCC11, 12, 13. In a previous study, we identified a natural mutant of the HBx gene with a deletion spanning 382 to 401 base pairs (HBxΔ127). The HBxΔ127 mutant promotes the activities of NF-κB, survivin, and human telomerase reverse transcriptase (hTERT) as well as the expression levels of c-Myc and proliferating cell nuclear antigen (PCNA) in normal liver cells14. Recently, we have reported that HBxΔ127 promotes hepatoma cell proliferation through upregulation of 5-lipoxygenases (5-LOX) and fatty acid synthase (FAS)15. The role of HBxΔ127 in the promotion of cell migration, however, remains unclear.
Osteopontin (OPN), a secreted phosphoprotein, was originally characterized in malignant-transformed mammalian epithelial cells16. Recent studies have demonstrated that OPN is associated with tumor metastasis, overexpressed in many tumor tissues, and correlated to metastatic tissues17, 18. OPN regulation in tumor metastasis involves multiple pathways, including AP-1, Myc, v-Src, Runx/CBF, TGF-B/BMPs/Smad/Hox, and Wnt/β–catenin/APC/GSK–3β/Tcf-419. Importantly, OPN is overexpressed in HCC tissues involving HCC invasion and metastasis18, 20. Xe H et al reported that OPN was positive in 39 of 72 (54.17%) HBV-related HCC tissue samples21. Due to its ubiquitous expression in many tumor types, OPN has been used as a biomarker of advanced disease and is considered a potential therapeutic target for the regulation of cancer metastasis22, 23. 5-LOX is one of three key enzymes associated with the metabolism of arachidonic acid to biologically active eicosanoids and is often overexpressed in multiple tumor types. It was also shown that 5-LOX expression increased in 8 of 8 human colon cancer surgical samples relative to normal colonic epithelium tissue24. In addition, our previous study revealed that 5-LOX was involved in the proliferation and migration of LM-MCF-7, a breast cancer cell line with high metastatic potential, and hepatoma HepG2 cells15, 25.
In the present study, we investigate a novel role for HBxΔ127 in the promotion of hepatoma cell migration and show that HBxΔ127 can activate OPN through 5-LOX in the process. Our findings provide new insight into the mechanism by which HBxΔ127 promotes migration in hepatoma cells.
Materials and methods
Plasmids, Reagents and siRNAs
The pSilencer3.0-X, pGL3-Basic, and pGL3-Control plasmids and the renilla luciferase reporter vector pRL-TK were described previously14, 26. The pGL3-OPN plasmid contained the firefly luciferase reporter and the full-length OPN promoter sequence. MK886, a specific inhibitor of 5-LOX, was purchased from Sigma-Aldrich (St Louis, MO, USA). The small interfering RNA (siRNA) targeting human OPN mRNA (targeting sequence: 5′-GCCACAAGCAGTCCAGATT-3′; D28759)27 and the negative control siRNA were designed and synthesized by RiboBio (Guangzhou, China).
Cell culture
Human hepatoblastoma HepG2 (ATCC HB 8065), human hepatocellular carcinoma H7402 (Purchased from People's Hospital, Beijing, China), HepG2-P/H7402-P (stably transfected with empty pCMV-Tag2B vector plasmid), HepG2-X/H7402-X (stably transfected with pCMV-X plasmid), and HepG2-X Δ127/H7402-X Δ127 (stably transfected with pCMV-X Δ127 plasmid) cell lines were used in this study and have been described previously15. All above cells lines were maintained in Dulbecco's modified Eagle's (DMEM) medium (Gibco, CA, USA) supplemented with heat inactivated 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
Wound healing assays
Cells were seeded in 6-well plates and grown to approximately 90% confluence before wounding with a 200 μL plastic tip across the monolayer. Debris was removed by washing three times with PBS, and then the cells were cultured with fresh medium containing 5% fetal bovine serum. Images were captured immediately after wounding and at 12, 24, and 36 (or 48) h post wounding28. The migration distance was calculated according to the formula: migration distance=(initial wound width – wound width at each time point)/2 (μm)29. Each experiment was performed in triplicate and repeated three times. The cells transfected with empty vector or cells transfected with control siRNA served as negative controls.
Construction of the human OPN promoter
The full-length promoter of the human OPN gene (from -2104 nt to +78 nt, including the first untranslated exon; GenBank S78410) was amplified using PCR primers (Table 1) based on the published sequence30. Human genomic DNA was used as a template. The full-length construct was cloned into the KpnI and XhoI sites of the firefly luciferase reporter plasmid pGL3-Basic vector (Promega, Madison, WI, USA).
RNA interference
HepG2-XΔ127 and H7402-X Δ127 cells were transfected with a pSilencer3.0-X vector, respectively, which expressed siRNA that targeted the HBxΔ127 mRNA (targeting to 271–290 nt of HBx mRNA), or the control siRNA26. Duplex siRNA targeting the human mRNA of OPN and control siRNA were introduced into HepG2-XΔ127 and H7402-XΔ127 cells according to the manufacturer's protocol. Each experiment included controls containing the transfection reagent with control siRNA. Transfected cells were subjected to luciferase reporter gene assays, RT-PCR and wound healing assays 48 h after transfection.
Treatment of tumor cells
HepG2-XΔ127 and H7402-XΔ127 cells were cultured in serum-free medium for 12 h. Then the engineered cells were treated with MK886 (5, 10, or 20 μmol/L) for 6 h as described previously15. The treated cells were subjected to luciferase reporter gene assays and RT-PCR. The examination of cytotoxicity mediated by MK886 was described previously15.
Luciferase reporter gene assays
Cells were seeded in 24-well plates (5×104 cells/well) and cultured in DMEM medium. The cells were transiently transfected with 0.3 μg of the luciferase reporter plasmids pGL3-OPN, pGL3-Basic, or pGL3-Control, respectively. The cells in each well were cotransfected with 50 ng renilla luciferase reporter vector pRL-TK. Cells were harvested after 48 h and lysed in 1×passive lysis buffer. The luciferase activity was determined using Dual-Luciferase Reporter® Assay System (Promega, Madison, WI, USA) on a luminometer (TD-20/20, Sunnyvale CA, USA) according to the manufacturer's protocol. The pGL3-Basic and pGL3-Control plasmid were used as negative and positive controls, respectively. Luciferase activity was normalized for transfection efficiency using the corresponding renilla luciferase activity by cotransfection with pRL-TK. All experiments were performed at least three times. The data of the luciferase reporter gene assays were analyzed by Student's t test to identify statistically significant differences.
RNA extraction and RT-PCR
Total cellular RNA was extracted using Trizol reagent (Invitrogen, CA, USA) from cells 48 h after treatment. cDNA was synthesized using the M-MLV RTase cDNA Synthesis Kit (Takara Bio Inc, Tokyo, Japan) according to the manufacturer's protocol. Synthesized cDNA was used as a template for PCR with Taq polymerase (94 °C for 3 min, 25 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, and 72 °C for 10 min). The PCR products were verified by 1% agarose gel electrophoresis with ethidium bromide and visualized with UV illumination. The primers used for PCR are listed in Table 1.
Western blot analysis
The cells were washed three times with ice-cold PBS and extracted directly in the lysis buffer (62.5 mmol/L Tris-HCl, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol). Equal amounts of protein (30 μg) were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a PVDF membrane for 90 min. The membrane was blocked in blocking buffer (PBS, 5% skim milk, 0.1% Tween 20) at room temperature for 2 h. Proteins on the membrane were hybridized at 4 °C overnight with the following the primary antibodies against HBx (1:1000 dilution, Abcom, Cambridge, UK) and β-actin (1:1000 dilution, Sigma-Aldrich, St Louis, MO, USA). The membranes were washed three times in PBS (0.1% Tween 20) and incubated for 1 h with an HRP-linked secondary antibody (Amersham Phamacia Biotech, NJ, USA). The membranes were then washed three times, and the protein bands were visualized by ECL reagent (Amersham Phamacia Biotech, NJ, USA).
Statistical analysis
All values are presented as means±SEM of at least three separate experiments. Data were analyzed by comparing two groups using Student's t test. P<0.05 was considered significant.
Results
HBxΔ127 strongly promotes hepatoma cell migration
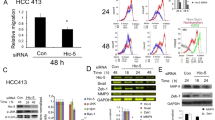
To investigate the function of HBxΔ127 in hepatoma cell migration, we examined HepG2-X and HepG2-XΔ127 cell migration ability. The wound healing assays showed that HepG2-XΔ127 cells exhibited a greater ability to repair the wound than did HepG2-X cells at 36 h post wounding (P<0.05, HepG2-XΔ127 vs HepG2-X and P<0.01, HepG2-X vs HepG2) (Figure 1A and 1B), suggesting that HBxΔ127 strongly promotes hepatoma cell migration relative to wild type HBx.
HBxΔ127 strongly promotes hepatoma cell migration. (A) The migration ability of HepG2-XΔ127 cells was examined with a wound healing assay. Images were taken at 0, 12, 24 and 36 h with a phase-contrast microscope (100×). Black arrows indicate the wound edge closure of monolayer cells. The results shown are representative of three independent experiments. (B) The average migration distance of the wound edge in three independent experiments. Compared with HepG2 cells, HepG2-X cells exhibited a greater ability to repair the wound (cP<0.01, HepG2-X cells vs HepG2 cells). Compared with HepG2-X cells, HepG2-XΔ127 cells exhibited significantly greater ability to repair the wound (eP<0.05, HepG2-XΔ127 cells vs HepG2-X cells).
HBxΔ127 enhances the promoter activity of OPN in hepatoma cells
Accordingly, OPN plays a crucial role in HCC invasion and metastasis. Thus, we hypothesized that OPN may be involved in HBxΔ127-mediated cell migration. To investigate the effect of HBxΔ127 on the promoter activity of OPN, we generated the pGL3-OPN plasmid, an OPN promoter luciferase reporter. The plasmid was constructed by first cloning the full-length promoter of the human OPN gene and then inserting the cloned promoter into the pGL3-Basic vector according to the published effective promoter sequence30 (Figure 2A). Additionally, transient transfection was performed in HepG2 and H7402 cell models using the pGL3-OPN plasmid. Luciferase reporter gene assays were used to verify that the OPN promoter was successfully cloned, with pGL3-Basic and pGL3-Control representing the negative and positive controls, respectively (P<0.01, pGL3-OPN vs pGL3-Basic) (Figure 2B and 2C). We then examined the effect of HBxΔ127 on the promoter activity of OPN using pGL3-OPN in luciferase reporter gene assays. Our data showed that the promoter activity of OPN was enhanced in HepG2-X/HepG2-XΔ127 and H7402-X/H7402-XΔ127 cells (P<0.01, HepG2-X vs HepG2 and HepG2-XΔ127 vs HepG2-X) (Figure 3A and 3B). In addition, RNA interference (RNAi) targeting HBxΔ127 mRNA mediated by pSilencer3.0-X (termed pSi-HBx) abolished the enhancement of the HBxΔ127-mediated OPN promoter activity in a dose-dependent manner (P<0.01, pSi-HBx vs control) (Figure 3C and 3D). These data suggest that HBx Δ127 strongly enhances the promoter activity of OPN in hepatoma cells.
Construction of the full-length promoter of the human OPN gene. (A) The PCR product of the full-length human OPN promoter is shown using the human genome as a template (B and C). The promoter activities of OPN were examined using a luciferase reporter gene assay in HepG2 and H7402 cells via co-transfection with pGL3-OPN plasmid and internal control vector (renilla luciferase reporter vector pRL-TK) (cP<0.01, pGL3-OPN vs pGL3-Basic). The results shown are representative of three independent experiments. The pGL3-Basic and pGL3-Control plasmids were the negative and positive controls, respectively.
HBxΔ127 strongly enhances the promoter activity of OPN in hepatoma cells. (A and B) A luciferase reporter gene assay showed that the promoter activities of OPN were significantly increased in HepG2-XΔ127 and H7402-XΔ127 cells compared to HepG2-X and H7402-X cells, respectively (fP<0.01, HepG2-XΔ127 vs HepG2-X and H7402-XΔ127 vs H7402-X), and increased in HepG2-X and H7402-X cells relative to HepG2 and H7402 cells, respectively (cP<0.01, HepG2-X vs HepG2 and H7402-X vs H7402). (C and D) The enhancement of OPN promoter activity was abolished by treatment with pSilence3.0-X (pSi-HBx) in HepG2-XΔ127 and H7402-XΔ127 cells in a dose-dependent manner (cP<0.01, vs untreated group).
HBxΔ127 increases the mRNA expression of OPN
To further investigate the influence of HBxΔ127 on OPN in hepatoma cells, we examined the mRNA level of OPN in HepG2-X/HepG2-XΔ127 and H7402-X/H7402-XΔ127 cells by RT-PCR. The data showed that the expression of OPN mRNA was significantly increased in HepG2-X/HepG2-XΔ127 and H7402-X/H7402-XΔ127 cells and that the expression level was higher in HepG2-XΔ127 and H7402-XΔ127 cells than in HepG2-X and H7402-X cells, respectively (Figure 4A and 4B). Meanwhile, the RNAi targeting HBxΔ127 mRNA was able to attenuate the upregulation of OPN at the mRNA level in a dose-dependent manner in HepG2-XΔ127 and H7402-XΔ127 cells (Figure 4A and 4B). Additionally, we found that HBxΔ127 mRNA expression was downregulated by treatment with pSi-HBx in a dose-dependent manner (Figure 4B), suggesting that HBxΔ127 strongly increases mRNA expression levels of OPN.
HBxΔ127 strongly increases mRNA expression levels of OPN. (A) The mRNA expression level of OPN was increased in HepG2-X/HepG2-XΔ127 and H7402-X/H7402-XΔ127 cells compared to HepG2 and H7402 cells, respectively. The expression was higher in the HepG2-XΔ127 and H7402-XΔ127 cells than in the HepaG2-X and H7402-X cells. GAPDH was the internal control. (B) The upregulation of HBxΔ127-mediated OPN mRNA expression was attenuated by treatment with pSilence3.0-X (pSi-HBx) in a dose-dependent manner. Meanwhile, the results confirm that the expression of HBxΔ127 mRNA is downregulated by pSi-HBx in the HepaG2-X/HepG2-XΔ127 cells. GAPDH was the internal control.
5-LOX is required for HBxΔ127 mediated upregulation of OPN
Our previous studies demonstrated that HBxΔ127 can upregulate the expression of 5-LOX15 and that 5-LOX is overexpressed in LM-MCF-7 and MDA-MB-231 cells involved in breast cancer metastasis25. Therefore, we hypothesized that 5-LOX may be involved in HBxΔ127-mediated upregulation of OPN. To test our hypothesis, we examined the effect of 5-LOX on the promoter activity and mRNA expression of OPN in HepG2-XΔ127 and H7402-XΔ127 cells. After treating the cells with MK886 (a specific 5-LOX inhibitor) for 6 h, we found that the enhancement of OPN promoter activity was abolished in HepG2-XΔ127 and H7402-XΔ127 cells in a dose-dependent manner (P<0.01 vs untreated group) (Figure 5A and 5B). Additionally, RT-PCR showed that MK886 attenuated the increased HBxΔ127-mediated expression of OPN mRNA in a dose-dependent manner in HepG2-XΔ127 and H7402-XΔ127 cells (Figure 5C). Thus, we conclude that 5-LOX is required for HBxΔ127-mediated upregulation of OPN.
5-LOX is required for the HBxΔ127-mediated upregulation of OPN. (A and B) Luciferase reporter gene assays showed that the promoter activities of OPN were decreased by treatment with the indicated doses of MK886 (a specific inhibitor of 5-LOX) for 6 h in HepG2-XΔ127 and H7402-XΔ127 cells (cP<0.01, vs untreated group). (C) RT-PCR showed that the expression levels of OPN mRNA were decreased by treatment with indicated doses of MK886 for 6 h in HepG2-XΔ127 and H7402-XΔ127 cells. GAPDH was the internal control. The mRNA levels of HBxΔ127 and 5-LOX were also utilized as controls.
OPN is involved in the HBxΔ127-mediated promotion of hepatoma cell migration
Next, we investigated the effect of OPN on the migration of HepG2-XΔ127 cells. The wound healing assays showed that the migration ability of HepG2-XΔ127 cells is suppressed by treatment with siRNA targeting OPN mRNA or pSi-HBx for 48 h (P<0.01, HepG2-XΔ127 cells treated with OPN siRNA or pSi-HBx vs control cells) (Figure 6A and 6B), suggesting that OPN is involved in the HBxΔ127-mediated promotion of hepatoma cell migration. The efficiency of OPN modulation by siRNAs was detected by RT-PCR, demonstrating that 100 nmol/L siRNA targeting OPN mRNA can significantly suppress the expression of OPN mRNA in HepG2-XΔ127 cells (Figure 6C). Finally, western blot analysis revealed that 2 μg pSi-HBx was able to significantly suppress the expression of HBxΔ127 in HepG2-XΔ127 cells (Figure 6D).
OPN is involved in HBxΔ127-mediated promotion of hepatoma cell migration. (A) Wound healing assays showed that the migration ability of HepG2-XΔ127 cells was suppressed by treatment with siRNA targeting OPN mRNA or pSi-HBx for 48 h. Images were taken from wound healing assays at 0, 12, 24, and 48 h in a phase-contrast microscope (100×). Black arrows indicate the wound edge closure of monolayer cells. The results shown are representative of three independent experiments. (B) The average migration distance of the wound edge in three independent experiments. The greater migration ability of HepG2-XΔ127 could be suppressed by knockdown of OPN or HBxΔ127 in a time-dependent manner (cP<0.01, vs untreated groups). (C) Efficiency of OPN modulation by treatment with 100 nmol/L siRNA targeting OPN mRNA was detected by RT-PCR. (D) The efficiency of HBxΔ127 modulation by treatment with 2 μg pSilence3.0-X (termed pSi-HBx) targeting HBxΔ127 mRNA was examined by Western blot analysis.
Discussion
HCC is one of the most common malignant tumors in Asia and has a very high mortality rate due to its high incidence of invasion and metastasis. HBV infection is associated with the development and metastasis of HCC31, 32. Many reports have shown that COOH-terminal truncated HBx plays a critical role in hepatocarcinogenesis through the regulation of cell proliferation, apoptosis, viability, and transformation11, 33. Liu et al reported that HBx30-20 and HBx30-40 mutants promote cell cycle progression from G0/G1 to S phase and may mechanistically increase invasive potential34. Our laboratory previously identified a natural mutant of HBx (HBxΔ127) that strongly promotes cell proliferation14, 15. Thus, in this study we attempted to discover a novel role for HBxΔ127 in the promotion of hepatoma cell migration.
Our findings show that HBxΔ127 has a strong ability to promote hepatoma HepG2 cell migration relative to wild-type HBx (Figure 1), suggesting that the natural mutant HBx plays an important role not only in the promotion of hepatoma cell proliferation but also in the promotion of hepatoma cell migration. Some reports have indicated that OPN is involved in tumor metastasis. OPN has been detected in tumor cells and the surrounding stroma of numerous human cancers, suggesting a correlation between high levels of OPN expression and malignant invasion20, 35. OPN mediates tumor metastasis by promoting tumor cell invasion and migration via the extracellular matrix, independent of cellular proliferation and cell-matrix adhesion. siRNAs targeting OPN mRNA significantly suppressed in vivo hepatic metastases, in vitro migration and invasion, and CT26 expression of matrix metalloproteinase-236. OPN function in vivo is multifaceted and involves multiple signaling pathways mediated by the αvβintegrin and CD44 receptors in the step-wise progression of metastasis37, 38. Thus, OPN is considered a biomarker for advanced disease as well as a potential therapeutic target in the regulation of cancer metastasis. We hypothesized, therefore, that OPN may be involved in the HBxΔ127-mediated promotion of cell migration. To probe this question, we examined the influence of HBxΔ127 on expression levels of OPN because the transcriptional regulation of the OPN promoter is a key event in the regulation of OPN expression and its synthesis. A number of studies involving critical mechanisms such as hormonal and growth factor stimulation, oncogene activation, and tumor development have focused on OPN gene expression at the mRNA level39. Therefore, we cloned the full-length promoter sequence of the human OPN gene (Figure 2) and observed the effect of HBxΔ127 on the promoter activity and mRNA expression of OPN in hepatoma cells. Our findings indicate that HBxΔ127 has a strong effect on the activation of OPN promoter activity and increases OPN expression at the mRNA level in hepatoma cells compared to the wild-type HBx gene. Our results also indicate that the upregulation of OPN can be abolished by treatment with HBxΔ127 RNAi (Figure 3 and 4), suggesting that HBxΔ127 activates OPN in hepatoma cells. In our experiments, we used two cell lines, HepG2 and H7402, as models. HepG2 is a human perpetual hepatoblastoma cell line, while H7402 is a human hepatocellular carcinoma cell line15, 40 used to replicate the experiments performed with the HepG2 cell line.
Next, we attempted to identify the mechanism of HBxΔ127-mediated upregulation of OPN. Our recent study indicated that HBxΔ127 can increase the expression of 5-LOX (a catalyzing enzyme of arachidonic acid) through phosphorylated extracellular signal-regulated protein kinases 1/2 (p-ERK1/2)15. Accordingly, 5-LOX is usually overexpressed during multistage tumor progression in many neoplastic disorders, including lung, breast, and pancreatic cancers24, 41. In addition, our laboratory found that 5-LOX was involved in the proliferation and migration of LM-MCF-7 (a breast cancer cell line with high metastatic potential25). Because OPN is an important mediator of tumor metastasis and correlates with tumor invasion, progression and metastasis in multiple cancers, we hypothesized that 5-LOX may be involved in increased HBxΔ127-mediated expression of OPN. Interestingly, we found that MK886 (a specific inhibitor of 5-LOX) was able to abolish the upregulation of OPN in HepG2-XΔ127 and H7402-XΔ127 cells (Figure 5), suggesting that 5-LOX is required for the upregulation of OPN. This finding is consistent with the report that 5-LOX is involved in tumor metastasis24. Furthermore, wound healing assays revealed that HepG2-XΔ127 cell migration can be significantly inhibited by treatment with siRNA targeting of OPN mRNA (or HBx mRNA) (Figure 6), further supporting the hypothesis that OPN is involved in HBxΔ127-mediated promotion of hepatoma cell migration.
Taken together, our data suggest that HBxΔ127 plays an important role not only in the promotion of hepatoma cell growth14, 15 but also in the promotion of hepatoma cell migration relative to wild-type HBx. HBxΔ127 strongly increased the expression of OPN through 5-LOX in the promotion of cell migration. This finding provides new insight into the mechanism of HBxΔ127-mediated promotion of hepatoma cell migration.
Author contribution
Xuan ZHANG, Prof Xiao-dong ZHANG, and Prof Li-hong YE designed the research; Xuan ZHANG performed the research; Xuan ZHANG analyzed the data and wrote the paper. Prof Xiao-dong ZHANG revised the paper.
References
Zhang XD, Zhang WY, Ye LH . Pathogenesis of hepatitis B virus infection. Future Virol 2006; 1: 637–47.
Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z . Genetic mechanisms of hepatocarcinogenesis. Oncogene 2002; 21: 2593–604.
Murakami S . Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol 2001; 36: 651–60.
Lee SG, Rho HM . Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene 2000; 19: 468–71.
Cougot D, Neuveut C, Buendia MA . HBV induced carcinogenesis. J Clin Virol 2005; 34 (Suppl 1): S75–S78.
Lara-Pezzi E, Gómez-Gaviro MV, Gálvez BG, Mira E, Iñiguez MA, Fresno M, et al. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J Clin Invest 2002; 110: 1831–8.
Chung TW, Lee YC, Kim CH . Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J 2004; 18: 1123–5.
Ou DP, Tao YM, Tang FQ, Yang LY . The hepatitis B virus X protein promotes hepatocellular carcinoma metastasis by upregulation of matrix metalloproteinases. Int J Cancer 2007; 120: 1208–14.
Lara-Pezzi E, Majano PL, Yáñez-Mó M, Gómez-Gonzalo M, Carretero M, Moreno-Otero R, et al. Effect of the hepatitis B virus HBx protein on integrin-mediated adhesion to and migration on extracellular matrix. J Hepatol 2001; 34: 409–15.
Lara-Pezzi E, Serrador JM, Montoya MC, Zamora D, Yáñez-Mó M, Carretero M, et al. The hepatitis B virus X protein (HBx) induces a migratory phenotype in a CD44-dependent manner: possible role of HBx in invasion and metastasis. Hepatology 2001; 33: 1270–81.
Liu XH, Wang L, Zhang SH, Lin J, Zhang SM, Feitelson MA, et al. Mutations in the carboxyl terminus of the X protein of Hepatitis B Virus regulate Wnt-5a expression in hepatoma Huh7 cells: cDNA microarray and proteomic analyses. Carcinogenesis 2008; 29: 1207–14.
Liu XH, Lin J, Zhang SH, Zhang SM, Feitelson MA, Gao HJ, et al. COOH-terminal deletion of HBx gene is a frequent event in HBV-associated hepatocellular carcinoma. World J Gastroenterol 2008; 14: 1346–52.
Iavarone M, Trabut JB, Delpuech O, Carnot F, Colombo M, Kremsdorf D, et al. Characterisation of hepatitis B virus X protein mutants in tumour and non-tumour liver cells using laser capture microdissection. J Hepatol 2003; 39: 253–61.
Zhang H, Shan CL, Li N, Zhang X, Zhang XZ, Xu FQ, et al. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol Sin 2008; 29: 473–80.
Wang Q, Zhang W, Liu Q, Zhang X, Lv N, Ye L, et al. A mutant of hepatitis b virus x protein (hbxδ127) promotes cell growth via a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia 2010; 12: 103–15.
Senger DR, Wirth DF, Hynes RO . Transformed mammalian cells secrete specific proteins and phosphoproteins. Cell 1979; 16: 885–93.
Rittling SR, Chambers AF . Role of osteopontin in tumor progression. Br J Cancer 2004; 90: 1877–81.
Pan HW, Ou YH, Peng SY, Liu SH, Lai PL, Lee PH, et al. Overexpression of osteopontin is associated with intrahepatic metastasis, early recurrence, and poorer prognosis of surgically resected hepatocellular carcinoma. Cancer 2003; 98: 119–27.
Wai PY, Kuo PC . Osteopontin: regulation in tumor metastasis. Cancer Metastasis Rev 2008; 27: 103–18.
Gotoh M, Sakamoto M, Kanetaka K, Chuuma M, Hirohashi S . Overexpression of osteopontin in hepatocellular carcinoma. Pathology International 2002; 52: 19–24.
Xie H, Song J, Du R, Liu K, Wang J, Tang H, et al. Prognostic significance of osteopontin in hepatitis B virus-related hepatocellular carcinoma. Dig Liver Dis 2007; 39: 167–72.
Yeatman TJ, Chambers AF . Osteopontin and colon cancer progression. Clin Exp Metastasis 2003; 20: 85–90.
Fedarko NS, Jain A, Karadag A, Van Eman MR, Fisher LW . Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clinical Cancer Research 2001; 7: 4060–6.
Melstrom LG, Bentrem DJ, Salabat MR, Kennedy TJ, Ding XZ, Strouch M, et al. Overexpression of 5-lipoxygenase in colon polyps and cancer and the effect of 5-LOX inhibitors in vitro and in a murine model. Clin Cancer Res 2008; 14: 6525–30.
You JC, Mi D, Zhou X, Qiao L, Zhang H, Zhang XD, et al. A positive feedback between activated erk and cox/lox maintains proliferation and migration of breast cancer cells. Endocrinology 2009; 150: 1607–17.
Zhang X, Dong N, Yin L, Cai N, Ma HT, You J, et al. Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J Med Virol 2005; 77: 374–81.
Lenga Y, Koh A, Perera AS, McCulloch CA, Sodek J, Zohar R . Osteopontin expression is required for myofibroblast differentiation. Circ Res 2008: 15; 102: 270–2.
Brown MD, Hart CA, Gazi E, Bagley S, Clarke NW . Promotion of prostatic metastatic migration towards human bone marrow stoma by Omega 6 and its inhibition by Omega 3 PUFAs. Br J Cancer 2006; 94: 842–53.
Wu KJ, Zeng J, Zhu GD, Zhang LL, Zhang D, Li L, et al. Silibinin inhibits prostate cancer invasion, motility and migration by suppressing vimentin and MMP-2 expression. Acta Pharmacol Sin 2009; 30: 1162–8.
Guo H, Marroquin CE, Wai PY, Kuo PC . Nitric Oxide-Dependent Osteopontin Expression Induces Metastatic Behavior in HepG2 Cells. Dig Dis Sci 2005; 50: 1288–98.
Zhang X, Zhang H, Ye L . Effects of hepatitis B virus X protein on the development of Liver cancer. J Lab Clin Med 2006; 147: 58–66.
Frati A, Salvati M, Giarnieri E, Santoro A, Rocchi G, Frati L . Brain metastasis from hepatocellular carcinoma associated with hepatitis B virus. J Exp Clin Cancer Res 2002; 21: 321–7.
Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res 2001; 61: 7803–10.
Liu X, Zhang S, Lin J, Zhang S, Feitelson MA, Gao H, et al. Biochem Biophys Hepatitis B virus X protein mutants exhibit distinct biological activities in hepatoma Huh7 cells. Res Commun 2008; 373: 643–7.
Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, et al. Osteopontin expression and distribution in human carcinomas. Am J Pathol 1994; 145: 610–23.
Wai PY, Mi Z, Guo H, Sarraf-Yazdi S, Gao C, Wei J, et al. Osteopontin silencing by small interfering RNA suppresses in vitro and in vivo CT26 murine colon adenocarcinoma metastasis. Carcinogenesis 2005; 26: 741–51.
Takahashi K, Takahashi F, Hirama M, Tanabe KK, Fukuchi Y . Restoration of CD44S in non-small cell lung cancer cells enhanced their susceptibility to the macrophage cytotoxicity. Lung Cancer 2003; 41: 145–53.
Bayless KJ, Salazar R, Davis GE . RGD-dependent vacuolation and lumen formation observed during endothelial cell morphogenesis in three-dimensional fibrin matrices involves the alpha(v)beta(3) and alpha(5)beta(1) integrins. Am J Pathol 2000; 156: 1673–83.
Denhardt DT, Noda M . Osteopontin expression and function: role in bone remodeling. J Cell Biochem Suppl 1998; 30–31: 92–102.
López-Terrada D, Cheung SW, Finegold MJ, Knowles BB . Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol 2009; 40: 1512–5.
Covey TM, Edes K, Fitzpatrick FA . Akt activation by arachidonic acid metabolism occurs via oxidation and inactivation of PTEN tumor suppressor. Oncogene 2007; 26: 5784–92.
Acknowledgements
This project was supported by grants from the National Basic Research Program of China (973 Program, No 2007CB914802, No 2007CB914804, No 2009CB521702) and the National Natural Science Foundation (No 30670959).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, X., Ye, Lh. & Zhang, Xd. A mutant of hepatitis B virus X protein (HBxΔ127) enhances hepatoma cell migration via osteopontin involving 5-lipoxygenase. Acta Pharmacol Sin 31, 593–600 (2010). https://doi.org/10.1038/aps.2010.36
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2010.36
Keywords
This article is cited by
-
Using 137Cs and 210Pbex to trace soil erosion rates for a small catchment in the mid-hills of Nepal
Journal of Soils and Sediments (2021)
-
miR-511 promotes the proliferation of human hepatoma cells by targeting the 3′UTR of B cell translocation gene 1 (BTG1) mRNA
Acta Pharmacologica Sinica (2017)
-
Hepatitis B virus X protein promotes human hepatoma cell growth via upregulation of transcription factor AP2α and sphingosine kinase 1
Acta Pharmacologica Sinica (2015)
-
Response of 210Pbex inventory to changes in soil erosion rates on uncultivated land
Chinese Science Bulletin (2013)