Abstract
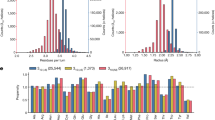
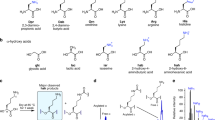
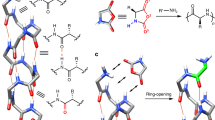
Proteins and RNA are unique among known polymers in their ability to adopt compact and well-defined folding patterns. These two biopolymers can perform complex chemical operations such as catalysis and highly selective recognition, and these functions are linked to folding in that the creation of an active site requires proper juxtaposition of reactive groups. So the development of new types of polymeric backbones with well-defined and predictable folding propensities ('foldamers') might lead to molecules with useful functions1,2. The first step in foldamer development is to identify synthetic oligomers with specific secondary structural preferences3–13. Whereas α-amino acids can adopt the well-known α-helical motif of proteins, it was shown recently11–13 that β-peptides3 constructed from carefully chosen β-amino acids can adopt a different, stable helical conformation defined by interwoven 14-membered-ring hydrogen bonds (a 14-helix; Fig. la). Here we report that β-amino acids can also be used to design β-peptides with a very different secondary structure, a 12-helix (Fig. la). This demonstrates that by altering the nature of β-peptide residues, one can exert rational control over the secondary structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Ball, P. Designing the Molecular World (Princeton Univ. Press, Princeton, 1994).
Lehn, J.-M. Supramolecular Chemistry: Concepts and Perspectives (VCH, Weinheim, 1995).
Dado, G. P. & Gellman, S. H. Intramolecular hydrogen bonding in derivatives of β-alanine and γ-amino butyric acid: model studies for the folding of unnatural polypeptide backbones. J. Am. Chem. Soc. 116, 1054–1062 (1994).
Hagihara, M., Anthony, N. J., Stout, T. J., Clardy, J. & Schreiber, S. L. Vinylogous polypeptides: an alternative peptide backbone. J. Am. Chem. Soc. 114, 6568–6570 (1992).
Eschenmoser, A. Chemistry of potentially prebiological natural products. Origins Life 24, 389–423 (1994).
Hamuro, Y., Geib, S. J. & Hamilton, A. H. Oligoanthranilamides. Non-peptide subunits that show formation of specific secondary structure. J. Am. Chem. Soc. 118, 7529–7541 (1996).
Gennari, C., Salom, B., Potenza, D. & Williams, A. Synthesis of sulfonamido-pseudopeptides: new chiral unnatural oligomers. Angew. Chem. Int. Edn Engl. 33, 2067–2069 (1994).
Lokey, R. S. & Iverson, B. L. Synthetic molecules that fold into a pleated secondary structure in solution. Nature 375, 303–305 (1995).
Smith, A. B. et al. De novo design, synthesis and X-ray crystal structures of pyrrolinone-based β-strand mimetics. J. Am. Chem. Soc. 116, 9947–9962 (1994).
Nowick, J. S., Mahrus, S., Smith, E. M. & Ziller, J. W. Triurea derivatives of diethylenetriamine as potential templates for the formation of artificial β-sheets. J. Am. Chem. Soc. 118, 1066–1072 (1996).
Seebach, D. et al. β-Peptides: synthesis by Arndt-Eistert homologation with concomitant peptide coupling. Structure determination by NMR and CD spectroscopy and by X-ray crystallography. Helical secondary structure of a β-hexapeptide in solution and its stability towards pepsin. Helv. Chim. Acta 79, 913–941 (1996).
Seebach, D. et al. Probing the helical secondary structure of short-chain β-peptides. Helv. Chim. Acta 79, 2043–2066 (1996).
Appella, D. H., Christianson, L. A., Karle, I. L., Powell, D. R. & Gellman, S. H. β-Peptide foldamers: robust helix formation in a new family of β-amino acid oligomers. J. Am. Chem. Soc. 118, 13071–13072 (1996).
Yuki, H., Okamoto, Y., Taketani, Y., Tsubota, T. & Marubayashi, Y. Poly(β-amino acid)s. IV. Synthesis and conformational properties of poly(α-isobutyl-L-aspartate). J. Polym. Sci. Polym. Chem. Edn 16, 2237–2251 (1978).
Fernández-Santin, J. M., Aymami, J., Rodríguez-Galan, A., Muñoz-Guerra, S. & Subirana, J. A. A pseudo α-helix from poly(α-isobutyl-L-aspartate), a nylon-3 derivative. Nature 311, 53–54 (1984).
Fernández-Santin, J. M. et al. Helical conformations in a polyamide of the nylon-3 family. Macromolecules 20, 62–68 (1987).
Bella, J., Alemán, C., Fernández-Santin, J. M., Alegre, C. & Subirana, J. A. Conformation of the helical polyamide poly(α-isobutyl-L-aspartate). Macromolecules 25, 5225–5230 (1992).
López-Carrasquero, F, Alemá, C. & Muñoz-Guerra, S. Conformational analysis of helical poly(β-L-aspartate)s by IR dichroism. Biopolymers 36, 263–271 (1995).
Herradon, B. & Seebach, D. Monoalkylation and dialkylation of derivatives of (IR, 2S)-2-hydro-xycyclopentanecarboxylic acid and (1R, 2S)-2-hydroxycyclohexanecarboxylic acid via bicyclic dioxanones: selective generation of three contiguous stereogenic centers on a cyclohexane ring. Helv. Chim. Acta 72, 690–714 (1989).
Tilley, J. W. et al. Analogs of Ac-CCK-7 incorporating dipeptide mimics in place of Met29–Gly29. J. Med. Chem. 35, 3774–3783 (1992).
Braunschweiler, L. & Ernst, R. R. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53, 521–528 (1983).
Bothner-By, A. A., Stephens, R. L., Lee, J., Warren, C. D. & Jeanloz, R. W. Structure determination of a tetrasaccharide: transient nuclear Overhauser effects in the rotating frame. J. Am. Chem. Soc. 106, 811–813 (1984).
Creighton, T. E. Proteins: Structures and Molecular Properties, 2nd Edn (Freeman, New York, 1993).
Xiong, H., Buckwalter, B. L., Shieh, H.-M. & Hecht, M. H. Periodicity of polar and nonpolar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. Proc. Natl Acad. Sci. USA 92, 6349–6353 (1995).
Schenck, H. L., Dado, G. P. & Gellman, S. H. Redox-triggered secondary structure changes in the aggregated states of a designed methionine-rich peptide. J. Am. Chem. Soc. 118, 12487–12494 (1996).
Barlow, D. J. & Thornton, J. M. Helix geometry in proteins. J. Mol. Biol. 201, 601–619 (1988).
Bacon, D. J. & Anderson, W. F. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graphics 6, 219–220 (1988).
Merrit, E. A. & Murphy, M. E. P. Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Cryst. D50, 869–873 (1994).
Kraulis, P. J. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950 (1991).
Brooks, B. R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Appella, D., Christianson, L., Klein, D. et al. Residue-based control of helix shape in β-peptide oligomers. Nature 387, 381–384 (1997). https://doi.org/10.1038/387381a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/387381a0
This article is cited by
-
Promiscuity mapping of the S100 protein family using a high-throughput holdup assay
Scientific Reports (2022)
-
Design of gel-to-sol UCST transition peptides by controlling polypeptide β-sheet nanostructures
Polymer Journal (2021)
-
Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic β-amino acids
Nature Chemistry (2020)
-
Enantiomeric glycosylated cationic block co-beta-peptides eradicate Staphylococcus aureus biofilms and antibiotic-tolerant persisters
Nature Communications (2019)
-
Highly functionalized cyclic β-amino acid moieties as promising scaffolds in peptide research and drug design
Amino Acids (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.