Abstract
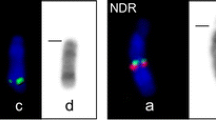
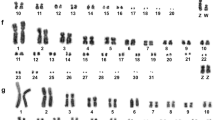
Q-band comparisons were made among representative species of the four genera of the tribe Bovini (Bos, Bison, Bubalus, Syncerus) as well as to selected outgroup taxa representing the remaining two tribes of the subfamily Bovinae (nilgai, Boselaphini; eland, Tragelphini), the Bovidae subfamily Caprinae (domestic sheep) and the family Cervidae (sika deer and white- tailed deer). Extensive autosomal arm homologies were noted, but relatively few derivative character states were shared. Focus was then made on variation of the sex chromosomes and the chromosomal distribution of nucleolar organizer regions (NORs). Bovine BAC clones were used in molecular cytogenetic analyses to decipher rearrangements of the sex chromosomes, and a pocket gopher 28s ribosomal probe was used to map the chromosomal locations of nucleolar organizing regions (NORs). Some of the more noteworthy conclusions drawn from the comparative analysis were that: 1. The Bovidae ancestral X chromosome was probably acrocentric and similar to acrocentric X chromosomes of the Bovinae; 2. The domestic sheep acrocentric X is probably a deriative character state that unites non-Bovinae subfamilies; 3. Bos and Bison are united within the tribe Bovini by the presence of shared derivative submetacentric X chromosomes; 4. Sika and white- tailed deer X chromosomes differ by inversion from X chromosomes of the Bovinae; 5. The Bovini ancestral Y chromosome was probably a small acrocentric; 6. Bos taurus, B. gaurus and B. banteng share derivative metacentric Y chromosomes; 7. Syncerus and Bubalus are united by the acquisition of X-specific repetitive DNA sequence on their Y chromosomes; 8. Bovinae and Cervidae X chromosome centromere position varies without concomitant change in locus order. Preliminary data indicate that a knowledge of the chromosomal distribution of NORs among the Bovidae will prove to be phylogenetically informative.
Similar content being viewed by others
References
Baker RJ, Davis SK, Bradely RD et al. (1989) Ribosomal-DNA, mitochondrial-DNA, chromosomal, and allozymic studies on a contact zone in the pocket gopher, Geomys. Evolution 43: 63–75.
Buckland RA, Evans HJ (1978) Cytogenetic aspects of phylogeny in the Bovidae. I. G-banding. Cytogenet Cell Genet 21: 42–63.
Cai L, Taylor JF, Wing RA et al. (1995) Construction and characterization of a bovine bacterial artificial chromosome library. Genomics 29: 413–425.
Committee for Standardized Karyotype of Bubalus bubalis (1994) Standardized karyotype of the river buffalo (Bubalus bubalis L., 2n = 50): report of the committee for the standardization of banded karyotypes of the river buffalo (Iannuzzi L, coordinator). Cytogenet Cell Genet 67: 102–113.
Davis SK (1986) Population Structure and Patterns of Speciation in Geomys (Rodentia: Geomyidae): An Analysis Using Mitochondrial and Ribosomal DNA. PhD Dissertation. Washington University, Saint Louis, MO, USA.
de Gortari MJ, Freking BA, Cuthbertson RP et al. (1998) A second generation linkage map of the sheep genome. Mammalian Genome 9: 204–209.
Di Meo GP, Perucatti A, Iannuzzi L et al. (1995) Constitutive heterochromatin polymorphism in river buffalo chromosomes. Caryologia 48: 137–145.
Effron M, Bogart MH, Kumamoto AT et al. (1976) Chromosome studies in the mammalian subfamily Antilopinae. Genetica 46: 419–444.
Eldridge FE (1985) Cytogenetics of Livestock. Westport, Connecticut: AVI Publishing Company, Inc.
Evans HJ, Buckland RA, Sumner AT (1973) Chromosome homology and heterochromatin in goat, sheep, and ox studied by banding techniques. Chromosoma 42: 383–402.
Fontana F, Rubini M (1990) Chromosomal evolution in Cervidae. Biosystems 24: 157–174.
Fries R, Popescu P (1999) Cytogenetics and physical chromosome maps. In: Fries R, Ruvinsky A, eds. The Genetics of Cattle. Wallington, UK: CABI Publishing.
Gallagher DS Jr, Womack JE (1992) Chromosome conservation in the Bovidae. J. Hered 83: 287–298.
Gallagher DS Jr, Derr JN, Womack JE (1994) Chromosome conservation among the advanced pecorans and determination of the primitive bovid karyotype. J Hered 85: 204–210.
Gallagher DS Jr, Davis SK, De Donato M et al. (1998a) A karyotypic analysis of the nilgai, Boselaphus tragocamelus (Artiodactyla: Bovidae). Chromosome Res 6: 505–513.
Gallagher DS Jr, Yang Y-P, Burzlaff JD et al. (1998b) Physical assignment of six type I anchor loci to bovine chromosome 19 by fluorescence in situ hybridization. Anim Genet 29: 130–134.
Groves CP (1969) Systematics of the anoa (Mammalia, Bovidae). Beaufortia, Series of Miscellaneous Publications, Zoological Museum, University of Amsterdam. 17(223): 1–12.
Iannuzzi L, Di Meo GP (1995) Chromosomal evolution in bovids: a comparison of cattle, sheep, and goat G-and R-banded chromosomes and cytogenetic divergences among cattle, goat and river buffalo sex chromosomes. Chromosome Res 3: 291–299.
Iannuzzi L, Di Meo GP, Perucatti A (1996) Identification of nucleolus organizer chromosomes and frequency of active NORs in river buffalo (Bubalus bubalis L.). Caryologia 49: 27–34.
ISCNDA (1990) International system for cytogenetic nomenclature of domestic animals. Cytogenet Cell Genet 53: 65–79.
Janis CM, Scott KM (1988) The interrelationships of higher ruminant families with special emphasis on the members of the Cervoidea. American Museum Novitates 2893: 1–85.
MacDonald DW (1987) The Encyclopedia of Mammals. New York: Facts on File.
Modi WS, Gallagher DS, Womack JE (1996) Evolutionary histories of highly repeated DNA families among the Artiodactyla (Mammalia). J Mol Evol 42: 337–349.
Petit P, Vermeesch JR, Marynen P et al. (1994) Comparative cytogenetic study in the subfamily Tragelaphinae. Proceedings of the 11th European Colloquium on Cytogenetics of Domestic Animals, pp. 109–113.
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83: 2934–2938.
Piumi F, Schibler L, Vaiman D et al. (1998) Comparative cytogenetic mapping reveals chromosome rearrangements between the X chromosomes of two closely related mammalian species (cattle and goat). Cytogenet Cell Genet 81: 36–41.
Popescu CP, Long S, Riggs P et al. (1996) Standardization of cattle chromosome nomenclature (1996). Report of the committee for the standardization of the cattle karyotype. Cytogenet Cell Genet 74: 259–261.
Qumsiyeh MB, Baker RJ (1988) Comparative cytogenetics and the determination of primitive karyotypes. Cytogenet Cell Genet 47: 100–103.
Reading Conference (1980) Proceedings of the first international conference for the standardization of banded karyotypes of domestic animals. Hereditas 92: 145–162.
Richardson AM (1997) Chromosome analysis. In: Barch MJ, ed. ACT Cytogenetics Laboratory Manual, 3rd edn. New York: Raven Press. pp. 481–526.
Robinson TJ, Harrison WR, Ponce de Leon A et al. (1997) X chromosome evolution in Suni and eland antelope: detection of homologous regions by fluorescence in situ hybridization and Gbanding. Cytogenet Cell Genet 77: 218–222.
Robinson TJ, Harrison WR, Ponce de Leon FA et al. (1998) A molecular cytogenetic analysis of X chromosome repatterning in the Bovidae: transposition, inversion and phylogenetic inference. Cytogenetic Cell Genet 80: 179–184.
Simpson GG (1945) The principles of classification, and a classification of mammals. Bull Am Mus Nat Hist 85: 1–350.
Solinas-Toldo S, Pienkowska A, Fries R et al. (1992) Localization of nucleolar organizer regions in farm animals by in situ hybridization method with a probe from human rRNA gene. Proceedings of the 10th European Colloquium on Cytogenetics of Domestic Animals, pp 232–236.
Vaughan TA (1986) Mammalogy, 2nd edn. New York: Saunders College Publishing.
Wilson DE, Reeder DM (1993) Mammal Species of the World, A Taxonomic and Geographic Reference, 2nd edn. Washington DC: Smithsonian Institution Press.
Wurster DH, Benirschke K (1968) Chromosome studies in the superfamily Bovoidea. Chromosoma 25: 152–171.
Yeh CC, Taylor JF, Gallagher DS et al. (1996) Genetic and physical mapping of the bovine X chromosome. Genomics 32: 245–252.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gallagher, D.S., Davis, S.K., De Donato, M. et al. A Molecular Cytogenetic Analysis of the Tribe Bovini (Artiodactyla: Bovidae: Bovinae) with an Emphasis on Sex Shromosome Morphology and NOR Distribution. Chromosome Res 7, 481–492 (1999). https://doi.org/10.1023/A:1009254014526
Issue Date:
DOI: https://doi.org/10.1023/A:1009254014526