Abstract
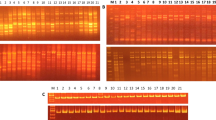
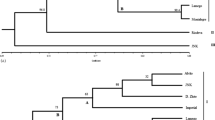
The present study, using RAPD analysis, was undertaken to characterize genetic variation in domesticated cowpea and its wild progenitor, as well as their relationships. The materials used consisted of 26 domesticated accessions, including accessions from each of the five cultivar-group, and 30 wild/weedy accessions, including accessions from West, East and southern Africa. A total of 28 primers generated 202 RAPD bands. One hundred and eight bands were polymorphic among the domesticated compared to 181 among wild/weedy cowpea accessions. Wild accessions were more diverse in East Africa, which is the likely area of origin of V. unguiculata var. spontanea. Var. spontanea is supposed to have spread westward and southward, with a loss of variability, loss counterbalanceed in southern Africa by introgressions with local perennial subspecies. Although the variabilty of domesticated cowpea was the highest ever recorded, cultivar-groups were poorly resolved, and several results obtained with isozyme data were not confirmed here. However primitive cultivars were more diverse than evolved cultivars, which still suggests two consecutive bottlenecks within domesticated cowpea evolution. As isozymes and AFLP markers, although with a larger number of markers, RAPD data confirmed the single domestication hypothesis, the gap between wild and domesticated cowpea, and the widespread introgression phenomena between wild and domesticated cowpea.
Similar content being viewed by others
References
Abo-Elwafa A., Murai K. and Shimada T. 1995. Intra-and inter-specific variations in Lens revealed by RAPD markers. Theor. Appl. Genet. 90: 335–340.
Ahmad M., McNeil D.L., Fautrier A.G., Armstrong K.F. and Paterson A.M. 1996. Genetic relationships in Lens species and parentage determination of their interspecific hybrids using RAPD markers. Theor. Appl. Genet. 92: 1091–1098.
Ahmad F. 1999. Random amplified polymorphic DNA (RAPD) analysis reveals genetic relationships among the annual Cicer species. Theor. Appl. Genet. 98: 657–663.
Akundabweni L.S.M. 1995. The potential of the arbitrarily primed polymerase chain reaction (AP-PCR) as a method to distinguish different genetic lines and strains in Vigna unguiculata (L.)Walp. S. Afr. J. Bot. 61: 5–8.
Amadou H.I., Bebeli P.J. and Kaltsikes P.J. 2001. Genetic diversity in Bambara groundnut (Vigna subterranea L.) germplasm re-vealed by RAPD markers. Genome 44: 995–999.
Banerjee H., Pai R.A. and Sharma R.P. 1999. Restriction fragment length polymorphism and random amplified polymorphic DNA analysis of chickpea accessions. Biol. Plant. 42: 197–208.
Baudoin J.P. and Maréchal R. 1985. Genetic diversity in Vigna. In: Singh S.R. and Rachie K.O. (eds), Cowpea research, production and utilization. John Wiley & Sons, Chichester, pp. 3–11.
Beebe S., Ochoa I., Skroch P., Nienhuis J. and Tivang J. 1995. Genetic diversity among common bean breeding lines developed for central America. Crop Sci. 35: 1178–1183.
Beebe S., Skroch P.W., Tohme J., Duque M.C., Pedraza F. and Nienhuis J. 2000. Structure of genetic diversity among common bean landraces of Middle American origin based on corre-spondence analysis of RAPD. Crop Sci. 40: 264–273.
Briand L., Brown A.E., Lenne J.M. and Teverson D.M. 1998. Random amplified polymorphic DNA variation within and among bean landrace mixtures (Phaseolus vulgaris L.) from Tanzania. Euphytica 102: 371–377.
Brown-Guedira G.L., Thompson J.A., Nelson R.L. and Warburton M.L. 2000. Evaluation of genetic diversity of soybean intro ductions and North American ancestors using RAPD and SSR markers. Crop Sci. 40: 815–823.
Cattan-Toupance I., Michalakis Y. and Neema C. 1998. Genetic structure of wild bean populations in their south-andean centre of origin. Theor. Appl. Genet. 96: 844–851.
Chevalier A. 1944. La dolique de Chine en Afrique. Rev. Bot. Appl. Agric. Trop. 24: 128–152.
Coulibaly S., Pasquet R.S., Papa R. and Gepts P. 2002. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L.Walp. reveals extensive gene flow between wild and domesticated types. Theor. Appl. Genet. 104: 358–366.
Dice L.R. 1945. Measures of the amount of ecologic association between species. Ecology. 26: 297–302.
Doldi M.L., Vollmann J. and Lelley T. 1997. Genetic diversity in soybean as determined by RAPD and microsatellite analysis. Plant Breed. 116: 331–335.
Doyle J.J. and Doyle J.L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15.
Duarte J.M., dos Santos J.B. and Melo L.C. 1999. Genetic diver-gence among common bean cultivars from different races based on RAPD markers. Genet. Mol. Biol. 22: 419–426.
Dwivedi S.L., Gurtu S., Chandra S., Yuejin W. and Nigam S.N. 2001. Assessment of genetic diversity among selected groundnut germplasm. I: RAPD analysis. Plant Breed. 120: 345–349.
Faris D.G. 1963. Evidence for the West African origin of Vigna sinensis (L.) Savi, PhD, University of California, Davis.
Ferguson M.E., Robertson L.D., Ford-Lloyd B.V. and Newbury H.J. 1998. Contrasting genetic variation amongst lentil landraces from different geographical origins. Euphytica 102: 265–273.
Fofana B., Vekemans X., Du Jardin P. and Baudoin J.P. 1997. Genetic diversity in Lima bean (Phaseolus lunatus L.) as revealed by RAPD markers. Euphytica 95: 157–165.
Fotso M., Azanza J.L., Pasquet R. and Raymond J. 1994. Molecular heterogeneity of cowpea (Vigna unguiculata, Fabaceae) seed storage proteins. Plant Syst. Evol. 191: 39–56.
Freyre R., Ríos R., Guzmán L., Debouck D. and Gepts P. 1996. Ecogeographic distribution of Phaseolus spp. (Fabaceae) in Bolivia. Econ Bot 50: 195–215.
Galvan M.Z., Aulicino M.B., Medina S.G. and Balatti P.A. 2001. Genetic diversity among Northwestern Argentinian cultivars of common bean (Phaseolus vulgaris L.) as revealed by RAPD markers. Genet. Resources Crop Evol. 48: 251–260.
Garba M. and Pasquet R.S. 1998. Isozyme polymorphism within section Reticulatae of genus Vigna (Tribe Phaseoleae: Fabaceae). Biochem. Syst. Ecol. 26: 297–308.
Gimenes M.A., Lopes C.R., Galgaro M.L., Valls J.E.M. and Koch-ert G. 2000. Genetic variation and phylogenetic relationships based on RAPD analysis in section Caulorrhizae, genus Arachis (Leguminosae). Euphytica 116: 187–195.
Haley S.D., Miklas P.N., Afanador L. and Kelly J.D. 1994. Random amplified polymorphic DNA (RAPD) marker variability be-tween and within gene pools of common bean. J. Amer. Soc. Hort. Sci. 119: 122–125.
Harlan J.R. 1971. Agricultural origins: centers and non centers. Science 174: 468–474.
Hilu K.W. and Stalker H.T. 1995. Genetic relationships between peanut and wild species of Arachis sect Arachis (Fabaceae): evidence from RAPDs. Plant Syst. Evol. 198: 167–178.
Johns M.A., Skroch P.W., Nienhuis J. and Hinrichsen P. 1997. Gene pool classification of common bean landraces from Chile based on RAPD and morphological data. Crop Sci. 37: 605–613.
Lanham P.G., Fennell S., Moss J.P. and Powell W. 1992. Detection of polymorphic loci in Arachis germplasm using random am-plified polymorphic DNAs. Genome 35: 885–889.
Li Z.L. and Nelson R.L. 2001. Genetic diversity among soybean accessions from three countries measured by RAPDs. Crop Sci. 41: 1337–1347.
Link W., Dixkens C., Singh M., Schwall M. and Melchinger A.E. 1995. Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor. Appl. Genet. 90: 27–32.
Liu C.J. 1996. Genetic diversity and relationships among Lablab purpureus genotypes evaluated using RAPD as markers. Euphytica 90: 115–119.
Maciel F.L., Gerald L.T.S. and Echeverrigaray S. 2001. Random amplified polymorphic DNA (RAPD) markers variability among cultivars and landraces of common beans (Phaseolus vulgaris L.) of south-Brazil. Euphytica 120: 257–263.
Maréchal R., Masherpa J.M. and Stainier F. 1978. Etude tax-onomique d'un groupe complexe d'espèces des genres Phaseolus et Vigna (Papilionaceae) sur la base des données morphologiques et polliniques, traitées par l'analyse informatique. Boissiera 28: 1–273.
Menéndez C.M., Hall A.E. and Gepts P. 1997. A genetic linkage map of cowpea (Vigna unguiculata) developed from a cross between two inbred domesticated lines. Theor. Appl. Genet. 95: 1210–1217.
Mienie C.M.S., Smit M.A. and Pretorius P.J. 1995. Use of random amplified polymorphic DNA for identification of South African soybean cultivars. Field. Crop Res. 43: 43–49.
Mignouna H.D., Ng N.Q., Ikea J. and Thottappilly G. 1998. Genetic diversity in cowpea as revealed by random amplified polymor phic DNA. J. Genet. Breed. 52: 151–159.
Mimura M., Yasuda K. and Yamaguchi H. 2000. RAPD variation in wild, weedy and cultivated azuki beans in Asia. Genet. Resour. Crop Evol. 47: 603–610.
Murdock G.P. 1959. Africa, Its Peoples and Their Culture History. McGraw Hill Book Company, New York.
Nei M. 1972. Genetic distance between populations. Am. Nat. 106: 283–292.
Nei M. 1973. Analysis of gene diversity in subdivided populations Proc. Natl. Acad. Sci. USA 70:, pp. 3321–3323.
Nei M. and Li W.H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases Proc. Natl. Acad. Sci. USA 76:, pp. 5269–5273.
Ng N.Q. 1995. Cowpea, Vigna unguiculata (Leguminosae-Papilionoideae). In: Smartt J. and Simmonds N.W. (eds), Evolu-tion of Crop Plants ed. 2. Longmans, New York, pp. 326–332.
Nienhuis J., Tivang J., Skroch P. and Dos Santos J.B. 1995. Genetic relationships among cultivars and landraces of lima bean (Phaseolus lunatus L.) as measured by RAPD markers. J. Amer. Soc. Hort. Sci. 120: 300–306.
Odeigah P.G.C. and Osanyinpeju A.O. 1996. Seed protein electrophoretic characterization of cowpea (Vigna unguiculata) germ-plasm from IITA bank. Genet. Resour. Crop Evol. 43: 485–491.
Padulosi S. 1993. Genetic diversity, taxonomy and ecogeographic survey of the wild relatives of cowpea (Vigna unguiculata (L.) Walpers.), PhD, Université catholique, Louvain La Neuve, Belgium.
Padulosi S. and Ng N.Q. 1997. Origin, taxonomy, and morphology of Vigna unguiculata (L.)Walp. In: Singh B.B., MohanRaj D.R., Dashiell K.E. and Jackai L.E.N. (eds), Advances in cowpea research. IITA-JIRCAS, Ibadan, pp. 1–12.
Paiva E., Vilarinhos A.D., Debarros E.G., Sediyama C.S. and Moreira M.A. 1994. Use of the random amplified polymorphic DNA technique to characterize soybean (Glycine max (L.) Mer-rill) genotypes. Rev. Bras. Genet. 17: 287–290.
Panella L. and Gepts P. 1992. Genetic relationship within Vigna unguiculata (L.) Walp. based on isozyme analyses. Genet. Re-sour. Crop Evol. 39: 71–88.
Panella L., Kami J. and Gepts P. 1993.Vignin diversity in wild and cultivated taxa of Vigna unguiculata (L.) Walp. (Fabaceae). Econ. Bot. 47: 371–386.
Pasquet R.S. 1993a. Classification infraspéécifique des formes spon-tanées de Vigna unguiculata (L.) Walp. à partir de données morphologiques. Bull. Jard. Bot. Nat. Belg. 62: 127–173.
Pasquet R.S. 1993b. Two new subspecies of Vigna unguiculata (L.) Walp. (Leguminosae: Papilionoideae). Kew Bull. 48: 805–806.
Pasquet R.S. 1993c. Variation at isozyme loci in wild Vigna un-guiculata (L.) Walp. (Fabaceae, Phaseoleae). Plant Syst. Evol. 186: 157–173.
Pasquet R.S. 1997. A new subspecies of Vigna unguiculata (Leguminosae-Papilionoideae). Kew Bull. 52: 840.
Pasquet R.S. 1998. Morphological study of cultivated cowpea Vigna unguiculata (L.) Walp. Importance of ovule number and definition of cv. gr. Melanophthalmus. Agronomie 18: 61–70.
Pasquet R.S. 1999. Genetic relationships among subspecies of Vigna unguiculata (L.) Walp. based on allozyme variation. Theor. Appl. Genet. 98: 1104–1119.
Pasquet R.S. 2000. Allozyme diversity of cultivated cowpea Vigna unguiculata (L.) Walp. Theor. Appl. Genet. 101: 211–219.
Pasquet R. 2001. Vigna Savi. In: Mackinder B., Pasquet R., Polhill R. and Verdcourt B. (eds), Flora Zambesiaca,volume part Phaseoleae. Royal Botanic Gardens, Kew, pp. 121–156.
Pasquet R.S. and Baudoin J.P. 2001. Cowpea. In: Charrier A., Jacquot M., Hamon S. and Nicolas D. (eds), Tropical Plant Breeding. Science publishers, Enfield, pp. 177–198.
Piper C.V. 1913. The wild prototype of cowpea. USDA Bureau of Plant Industry. Circular No 124. Miscellaneous papers. Washing-ton, Government Printing Office: 29–32.
Raina S.N., Rani V., Kojima T., Ogihara Y., Singh K.P. and Devarumath R.M. 2001. RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identifi-cation, and phylogenetic relationships in peanut (Arachis hypo-gaea) cultivars and wild species. Genome 44: 763–772.
Ratnaparkhe M.B., Gupta V.S., Venmurthy M.R. and Ranjekar P.K. 1995. Genetic fingerprinting of pigeonpea [Cajanus cajan (L.) Millsp.] and its wild relatives using RAPD markers. Theor. Appl. Genet. 91: 893–898.
Rawal K.M. 1975. Natural hybridization among wild, weedy and cultivated Vigna unguiculata (L.)Walp. Euphytica 24: 699–707.
Richard A. 1847. Tentamen florae abyssinicae, volumen primum. Arthus Bertrand, Paris.
Rohlf F.J. 1993. NTSYS-pc Numerical taxonomy and multivariate analysis system, version 1.80. Exeter software. Applied Bio statistics Inc., New York.
Saitou N. and Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Samec P. and Nasinec V. 1995. Detection of DNA polymorphism among pea cultivars using RAPD tehnique. Biol. Plant. 37: 321–327.
Santalla M., Power J.B. and Davey M.R. 1998. Genetic diversity in mung bean germplasm revealed by RAPD markers. Plant Breed. 117: 473–478.
Sharma S.K., Dawson I.K. and Waugh R. 1995. Relationships among cultivated and wild lentils revealed by RAPD analysis. Theor. Appl. Genet. 91: 647–654.
Singh B.B., Chambliss O.L. and Sharma B. 1997. Recent advances in cowpea breeding. In: Singh B.B., MohanRaj D.R., Dashiell K.E. and Jackai L.E.N. (eds), Advances in cowpea research. IITA-JIRCAS, Ibadan, pp. 30–49.
Skroch P.W. and Nienhuis J. 1995. Qualitative and quantitative characterization of RAPD variation among snap bean (Phaseolus vulgaris) genotypes. Theor. Appl. Genet. 91: 1078–1085.
Sonnante G. and Pignone D. 2001. Assessment of genetic variation in a collection of lentil using molecular tools. Euphytica 120: 301–307.
Steele W.M. 1972. Cowpeas in Nigeria, PhD, University of Read-ing, UK.
Steele W.M. 1976. Cowpeas, Vigna unguiculata (Leguminosaeguiculata Papillionatae). In: Simmonds N.W. (ed.), Evolution of Crop Plants. Longman, London, pp. 183–185.
Subramanian V., Gurtu S., Nageswara Rao R.C. and Nigam S.N. 2000. Identification of DNA polymorphism in cultivated ground-nut using random amplified polymorphic DNA (RAPD) assay. Genome 43: 656–660.
Thompson J.A., Nelson R.L. and Vodkin L.O. 1998. Identification of diverse soybean germplasm using RAPD markers. Crop Sci. 38: 1348–1355.
Tosti N. and Negri V. 2002. Efficiency of three PCR-based markers in assessing genetic variation among cowpea (Vigna un subsp. unguiculata) landraces. Genome 45: 268–275.
Vaillancourt R.E. and Weeden N.F. 1992. Chloroplast DNA poly-morphism suggests a Nigerian center of domestication for the cowpea, Vigna unguiculata (Leguminosae). Am. J. Bot. 79: 1194–1199.
Vaillancourt R.E., Weeden N.F. and Barnard J. 1993. Isozyme diversity in the cowpea species complex. Crop Sci. 33: 606–613.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ba, F.S., Pasquet, R.S. & Gepts, P. Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genetic Resources and Crop Evolution 51, 539–550 (2004). https://doi.org/10.1023/B:GRES.0000024158.83190.4e
Issue Date:
DOI: https://doi.org/10.1023/B:GRES.0000024158.83190.4e