Abstract
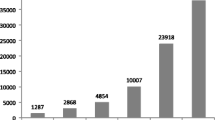
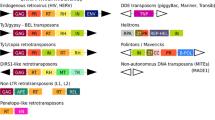
Interspersed repetitive sequences are major components of eukaryotic genomes. Repetitive elements comprise about 50% of the mammalian genome. They interact with the whole genome and influence its evolution. Repetitive elements may serve as recombination hot spots or acquire specific cellular functions such as RNA transcription control or become part of protein coding regions. The latter is a subject of presented analysis. We searched all currently available vertebrate protein sequences, including human proteome complement for the presence of transposable elements. It appears that insertion of TE-cassettes into open reading frames is a general phenomena. They can be found in all vertebrate lineages and originate in all types of transposable elements. It seems that genomes use those cassettes as ‘ready to use’ motifs in their evolutionary experiments. Most of TE-cassettes are used to create alternative forms of a message and usually the other form, without TE-cassette, is expressed in a cell. Tables listing vertebrate messages with TE-cassettes are available at http://warta.bio.psu.edu/ScrapYard/.
Similar content being viewed by others
References
Brosius, J., 1991. Retroposons-seeds of evolution. Science 251(4995): 753.
Cairns, J.S., J.M. Curtsinger et al., 1985. Sequence polymorphism of HLA DR beta 1 alleles relating to T-cell-recognized determinants. Nature 317(6033): 166-168.
Cairns, J.S., C.A. Dahl et al., 1988. Identification of a novel DR beta cDNA clone. Nucl Acids Res 16(19): 9353.
Caras, I.W., M.A. Davitz et al., 1987. Cloning of decay-accelerating factor suggests novel use of splicing to generate two proteins. Nature 325(6104): 545-549.
Claverie, J.M., 1992. Identifying coding exons by similarity search: alu-derived and other potentially misleading protein sequences. Genomics 12(4): 838-841.
Doolittle, W.F. & C. Sapienza, 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284(5757): 601-603.
Finnegan, D.J., 1989. Eukaryotic transposable elements and genome evolution. Trends Genet. 5(4): 103-107.
Gregersen, P.K., M. Shen et al., 1986. Molecular diversity of HLADR4 haplotypes. Proc Natl Acad Sci USA 83(8): 2642-2646.
Hickey, D.A., 1982. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics 101(3-4): 519-531.
Jurka, J., 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16(9): 418-420.
Maka?owska, I., R. Sood et al., 2002. Identification of six novel genes by experimental validation of GeneMachine predicted genes. Gene in press.
Maka?owski, W., 1995. pp. 86-104 in SINEs as a genomic scrap yard: an essay on genomic evolution, The Impact of Short Interspersed Elements (SINEs) on the Hpst Genome edited by R.J. Maraia. R.G. Landes Company, Austin.
Maka?owski, W., 2000. Genomic scrap yard: how genomes utilize all that junk. Gene 259(1-2): 61-67.
Maka?owski, W., G.A. Mitchell et al., 1994. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet 10(6): 188-193.
Orgel, L.E. & F.H. Crick, 1980. Selfish DNA: the ultimate parasite. Nature 284(5757): 604-607.
Pruitt, K.D. & D.R. Maglott, 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucl Acids Res 29(1): 137-140.
The Gene Ontology Consortium, 2001. Creating the gene ontology resource: design and implementation. Genome Res 11(8): 1425-1433.
Tugendreich, S., Q. Feng et al., 1994. Alu sequences in RMSA-1 protein? Nature 370(6485): 106.
Zhang, M.Q., 1998. Statistical features of human exons and their flanking regions. Hum Mol Genet 7(5): 919-932.
Zietkiewicz, E., W. Maka?owski et al., 1994. Phylogenetic analysis of a reported complementary DNA sequence. Science 265(5175): 1110-1111.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lorenc, A., Makałowski, W. Transposable Elements and Vertebrate Protein Diversity. Genetica 118, 183–191 (2003). https://doi.org/10.1023/A:1024105726123
Issue Date:
DOI: https://doi.org/10.1023/A:1024105726123