Abstract
Over the last decade the modeling and the storage of biological data has been a topic of wide interest for scientists dealing with biological and biomedical research. Currently most data is still stored in text files which leads to data redundancies and file chaos.
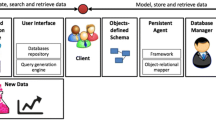
In this paper we show how to use relational modeling techniques and relational database technology for modeling and storing biological sequence data, i.e. for data maintained in collections like EMBL or SWISS-PROT to better serve the needs for these application domains.
For this reason we propose a two step approach. First, we model the structure (and therefore the meaning of the) data using an Entity-Relationship approach. The ER model leads to a clean design of a relational database schema for storing and retrieving the DNA and protein data extracted from various sources. Our approach provides the clean basis for building complex biological applications that are more amenable to changes and software ports than their file-base counterparts.
Similar content being viewed by others
REFERENCES
Aho, A. V., B.W. Kernighan and P.J. Weinberger (1988). The awk Programming Language. Addison-Wesley, Boston.
Bairoch, A. and R. Apweiler. (1999). The SWISS-PROT protein sequence databank and ist supplement TrEMBL in 1999. Nucleic Acids Research 27: 49-54.
Barker, W.C., J.S. Garavelli, P.B. McGarvey, C.R. Marzec, B.C. Orcutt, G.Y. Srinivasarao, L.S. Yeh, R.S. Ledley, H.W. Mewes, F. Pfeiffer, A. Tsugita and C. Wu. (1999). The PIR-International Protein sequence database. Nucleic Acids Research 27: 39-43.
Benson, D.A., M.S. Boguski, D.J. Lipman, J. Ostell, B.F. Ouellette, B.A. Rapp and D.L. Wheeler (1999). GenBank. Nucleic Acids Research 27: 12-17.
Bergholz, A., S. Heymann, J.A. Schenk and J.C. Freytag (1997). Sequence comparison using a relational database approach. Proceedings of International Database and Engineering and Applications Symposium 126-131.
Cariello, N. F., G.R. Douglas, M.J. Dycaico, N.J. Gorelick, G.S. Provost and T. Soussi (1997). Databases and software for the analysis of mutations in the human p53 gene, human hprt gene and both the lacI and lac/ gene in transgenic rodents. Nucleic Acids Research 25: 136-137.
Chen, P. P.-S. (1976). The Entity-Relationship-Model — Toward a Unified View of Data. ACM Transactions on Database Systems 1: 9-36.
Contrino, S. (2000). SWISS-PROT goes to Oracle http://www.ebi.ac.uk/~contrino/sp/
Date, C.J. (1995). An Introduction To Database Systems. The System Programming Series, 6th edition. Addison-Wesley, Boston.
EMBL Nucleotide Sequence Database Release Notes (Release 55, 1998). Available from ftp.ebi.ac.uk
Kabat, E. A., T.T. Wu, H.M. Perry, K.S. Gottesman and C. Foeller (1991). Sequences of Proteins of Immunological Interest. National Institutes of Health Publications No. 91: 3242.
Keen G., J. Burton, D. Crowley, E. Dickinson, A. Espinosa-Lujan, E. Franks, C. Harger, M. Manning, S. March, M. McLeod, J. O'Neill, A. Power, M. Pumilia, R. Reinert, D. Rider, J. Rohrlich, J. Schwertfeger, L. Smyth, N. Thayer, C. Troup and C. Fields (1996). The Genome Sequence DataBase (GSDB): meeting the challenge of genome sequencing. Nucleic Acids Research 24: 13-16.
Letovsky, S.I., R.W. Cottingham, C.J. Porter and P.W. Li (1998). GDB: the human genome database. Nucleic Acids Research 26: 94-99.
Moore, J., A. Engelberg and A. Bairoch (1988). Using PC/Gene for protein and nucleic acid analysis. Biotechniques 6: 566-572.
Ritter, O. (1994). The integrated genomic database. Computational Methods in Genome Research: 57-73.
Senger, M., K.H. Glatting, O. Ritter and S. Suhai (1995). X-HUSAR, an X-based graphical interface for the analysis of genome sequences. Computational Methods and Programs in Biomedicine 46: 131-141.
Stoesser, G., M.A. Tuli, P. Lopez and P. Sterk (1999). The EMBL Nucleotide sequence database. Nucleic Acids Research 27: 18-24.
Teorey, T. J., D. Yang and J.P. Fry (1986). A Logical Design Methodology for Relational Databases Using the Extended Entity-Relationship Model. ACM Computing Surveys 18: 197-222.
Thierry-Mieg, J. and R. Durbin (1992). Syntactic definitions for the ACEDB data base. Technical Report MRC-LMB xx.92.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bergholz, A., Heymann, S., Schenk, J.A. et al. Biological Sequences Integrated: A Relational Database Approach. Acta Biotheor 49, 145–159 (2001). https://doi.org/10.1023/A:1011958524279
Issue Date:
DOI: https://doi.org/10.1023/A:1011958524279