Abstract
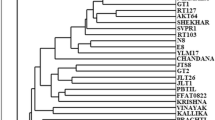
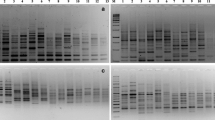
Fifty cassava clones were studied using RAPD technique. They included landraces from the Wenchi, Nkoranza, Dormaa Ahenkoro and Asonafo districts of the Brong Ahafo region of Ghana and three improved varieties. Genetic diversity of these genotypes was studied using four primers, OPK-01, OPR-02, OPR-09 and OPJ-14. A total of 41 different bands were detected. Levels of polymorphic fragments detected by the four primers ranged from 90% to 100%. By pooling bands from individual accessions together, mean number of fragments per accession per primer ranged from 5.50±1.04 for the Improved cultivars to 7.00±0.71 for populations of landraces from Dormaa. Mean frequencies of fragments not detected by the primers for the accessions were 0.524±0.12, 0.460±0.12, 0.561±0.12 and 0.523±0.12 for landraces from Wenchi, Nkoranza, Dormaa Ahenkro, Asonafo and the Improved varieties, respectively. The grand mean frequency of individuals showing fragments not present in populations was 0.522±0.10. Genetic diversity estimates ranged from 0.290 to 0.425 (mean 0.352±0.05) for primer OPK-01, 0.001 to 0.381 (mean 0.309±0.06) for primer OPR-02, 0.335 to 0.344 (mean 0.283±0.04) for primer OPR-09 and 0.152 to 0.352 (mean 0.261±0.07) for primer OPJ-14. Within the accessions mean gene diversity estimates were 0.316±0.03, 0.293±0.09, 0.331±0.02, 0.322±0.07 and 0.247±0.03 for accessions from Wenchi, Nkoranza, Dormaa Ahenkro, Asonafo districts and the Improved varieties, respectively. Interpopulational genetic divergence ranged from 0.069 to 0.203 (mean 0.119±0.04). Rate of nucleotide substitution among the landraces was 9.8 per cent per site per year, while that for the Improved varieties was 15 per cent.
Similar content being viewed by others
References
Asemota, H.N., 1995. A fast, simple and efficient miniscale method for the preparation of DNA from tissues of yam (Dioscorea spp.). Plant Mol Biol Rep 13: 214-218.
Adams, R.P. & T. Demeke, 1993. Systematic relationships in Juniperus based on random amplified polymorphic DNA (RAPDs). Taxon 42: 553-571.
Demeke, T., R.P. Adams & R. Chibbar, 1992. Potential use of random amplified polymorphic DNA (RAPDs): A case study with Brassica. Theor. Appl Genet 84: 990-994.
Doyle, D.J. & J.L. Doyle, 1990. A rapid total DNA preparation procedure for fresh plant tissue. Focus 12: 13-15.
Ellsworth, D.L., K.D. Rittenhouse & R.L. Honeycutt, 1993. Artifactual variation in randomly amplified polymorphic DNA banding patterns. Biotechnique 14: 214-217.
Hu, J. & C.F. Quiros, 1991. Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Rep 10: 505-511.
Isabel, N., J. Belieu & J. Bousquet, 1995. Complete congruence between gene diversity etimates derived from genotypic data at enzyme and random amplified polymorphic DNA loci in black spruce. Proc Natl Acad Sci USA 92: 6369-6373.
Kazan, K. & F.J. Muchlbauer, 1993. Allozyme variation and phylogeny in annual species of Cicer (Leguminosae). Plant Syst Evo 175: 11-21.
Koller, B., B. Lehmann, J.M. McDermott & C. Gessler, 1993. Identification of apple cultivars using RAPD markers. Theor Appl Genet 85: 901-904.
Marmey, P., J.R. Beeching, S. Hamon & A. Charrier, 1994. Evaluation of cassava (Manihot esculenta Crantz) germplasm collection using RAPD markers. Euphytica 74: 203-209.
Mayer, M.S., A. Tullu, C.J. Simon, J. Kumar, W.J. Kaiser, J.M. Kraft & F.J. Muelbauer, 1997. Development of a DNAmarker for Fusarium wilt resistance in chickpea. Crop Sci 37: 1625-1629.
Muralidhavan, K. & E.K. Wakeband, 1993. Concentration of primer and template qualitatively affects products in random-amplified polymorphic DNA PCR. Biotechnique 14: 362-364.
Nei, M. & J.C. Miller, 1990. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125: 873-879.
Roa, A.C., M.M. Maya, M.C. Duque, J. Tohme, A.C. Allem & M.W. Bonierbale, 1997. AFLP analysis of relationships among cassava and other Manihot species. Theor Appl Genet 95: 741-750.
Sneath, P.H.A. & R.R. Sokal, 1973. Numerical Taxonomy. The Principle and Practice of Numerical Classification, W.H. Freeman and Company. San Francisco.
Williams, J.G.K., A.R. Kubelik, K.J. Livak, J.A. Ratalski & S.V. Tingey, 1990. DNA polymorphims amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531-6535.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Asante, I.K., Offei, S.K. RAPD-based genetic diversity study of fifty cassava (Manihot esculenta Crantz) genotypes. Euphytica 131, 113–119 (2003). https://doi.org/10.1023/A:1023056313776
Issue Date:
DOI: https://doi.org/10.1023/A:1023056313776