Abstract
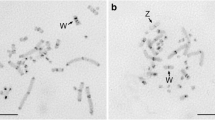
A novel repeated sequence of chaffinch (Fringilla coelebs) designated as GS was isolated from genomic DNA after in vitro amplification of satellite DNA sequences using GSP–PCR technique. The proportion of this repeat in the chaffinch genome constitutes about 0.2%. Monomers are 176 to 199 bp in size and contain a short cluster of the TTAGGG telomeric tandem repeat. The oligomer of the telomeric hexanucleotide is flanked by the sequences that are significantly different in different monomers. The GS sequences are organized as tandemly repeated units and located in a number of chromomycin-positive blocks on the long arms of macrochromosomes 1, 2, 3, 5, and 6, as well as on several microchromosomes. The sequences homologous to the GS satellite of chaffinch were not found in the genomes of redwing (Turdus iliacus) and house sparrow (Passer domesticus).
Similar content being viewed by others
REFERENCES
Mizuno, S. and Macgregor, H., The ZW Lampbrush Chromosomes of Birds: A Unique Opportunity to Look at the Molecular Cytogenetics of Sex Chromosomes, Cytogenet. Cell Genet., 1998, vol. 80, pp. 149-157.
Solovei, I.V., Joffe, B.I., Gaginskaya, E.R., and Macgregor, H.C., Transcription on Lampbrush Chromosomes of a Centromerically Localized Highly Repeated DNA in Pigeon (Columba) Relates to Sequence Arrangement, Chromosome Res., 1996, vol. 4, pp. 588-603.
Saifitdinova, A.F., Derjusheva, S.E., Malykh, A.G., et al., Centromeric Tandem Repeat from the Chaffinch Genome: Isolation and Molecular Characterization, Genome, 2001, vol. 44, pp. 96-103.
Madsen, C.S., Brooks, J.E., de Kloet, E., and de Kloet, S.R., Sequence Conservation of an Avian Centromeric Repeated DNA Component, Genome, 1994, vol. 37, pp. 351-355.
Matzke, M.A., Varga, F., Berger, H., et al., A 41-42 bp Tandemly Repeated Sequence Isolated from Nuclear Envelopes of Chicken Erythrocytes Is Located Predominantly on Microchromosomes, Chromosoma 1990, vol. 99, pp. 131-137.
Matzke, A.J.M., Varga, F., Gruendler, P., et al., Characterization of a New Repetitive Sequence That Is Enriched on Microchromosomes of Turkey, Chromosoma 1992, vol. 102, pp. 9-14.
Tanaka, K., Suzuki, T., Nojiri, T., et al., Characterization and Chromosomal Distribution of a Novel Satellite DNA Sequence of Japanese Quail (Coturnix coturnix japonica), J. Hered., 2000, vol. 91, pp. 412-415.
Evteev, A.V., Andreeva, T.F., and Gaginskaya, E.R., Transcriptional Analysis of the CNM Subtelomeric Repetitive Sequence in Chicken Microbivalents during Oogenesis, Tsitologiya, 2000, vol. 42, pp. 278-279.
Zhu, X., Skinner, J., and Burgoyne, L.A., The Macrosatellites of the Toulouse Goose: The Major Tandemly Repetitive DNA in the Toulouse Goose Genome, Genome, 1990, vol. 33, pp. 641-645.
Zhu, X., Skinner, J., and Burgoyne, L.A., The Interspersed DNA Repeat Conserved in Water Birds' Genomes, Genome, 1991, vol. 34, pp. 493-494.
Zhu, X.Y., Testori, A., Oh, S., et al., Avian Tandem Repeats: Nuclear Protein-Binding Sites and the Origins of Short Tandem Repetitive DNAs, Biochem. Int., 1991, vol. 25, pp. 191-198.
Zhu, X., Testori, A., Oh, S., et al., Avian Species-Specific Tandem Repeats Contain Nuclear Protein-Binding Sites, Biochem. Biophys. Res. Commun., 1992, vol. 182, pp. 447-451.
Keyser, C., Montagnon, D., Schlee, M., et al., First Isolation of Tandemly Repeated DNA Sequences in New-World Vultures and Phylogenetic Implications, Genome 1996, vol. 39, pp. 31-39.
Buntjer, J.B. and Lenstra, J.A., Self-Amplification of Satellite DNA in vitro, Genome, 1998, vol. 41, pp. 429-434.
Tiersch, T.R. and Wachtel, S.S., On the Evolution of Genome Size of Birds, J. Hered. 1991, vol. 82, pp. 363-368.
Malykh, A.G., Zhurov, V.G., Saifitdinova, A.F., et al., Structural and Functional Characterization of a Centro-meric Highly Repetitive DNA Sequence of Fringilla coelebs L. (Aves: Passeriformes), Mol. Biol. (Moscow), 2001, vol. 35, no. 3, pp. 331-335.
Altschul, S.F., Madden, T.L., Schaffer, A.A., et al., Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs, Nucleic Acids Res. 1997, vol. 25, pp. 3389-3402.
Lichter, P. and Cremer, T., Chromosome Analysis by Non-Isotopic in situ Hybridization, Human Cytogenet-ics: A Practical Approach, Oxford: IRL, 1992, pp. 157-192.
Toth, G., Gaspari, Z., and Jurka, J., Microsatellites in Different Eukaryotic Genomes: Survey and Analysis, Genome Res., 2000, vol. 10, pp. 967-981.
Garrido-Ramos, M.A., de la Herran, R., Ruiz Rejon, C., and Ruiz Rejon, M., A Satellite DNA of the Sparidae Family (Pisces, Perciformes) Associated with Telomeric Sequences, Cytogenet. Cell Genet. 1998, vol. 83, pp. 3-9.
Rodionov, A.V., GT-Reach Chromomycin-Positive (CMA/DA) Blocks in the Karyotypes of Chicken Gallus gallus domesticus: An Ideogram and Intraspecific Pat-tern Variation, Genetika (Moscow), 1998, vol. 34, no. 11, pp. 1523-1527.
Meyne, J., Baker, R.J., Hobart, H.H., et al., Distribution of Non-Telomeric Sites of the (TTAGGG)n Telomeric Sequence in Vertebrate Chromosomes, Chromosoma, 1990, vol. 99, pp. 3-10.
Fagundes, V., Vianna-Morgante, A.M., and Yonenaga-Yassuda, T., Telomeric Sequence Localization and G-Banding Patterns in the Identification of a Polymorphic Chromosomal Rearrangement in the Rodent Akodon cursor (2n = 14, 15, and 16), Chromosome Res., 1997, vol. 5, pp. 228-232.
Garagna, S., Ronchetti, E., Mascheretti, S., et al., Non-Telomeric Chromosome Localization of (TTAGGG)n Repeats in the Genus Eulemur, Chromosome Res., 1997, vol. 5, pp. 487-491.
Ijdo, J.W., Baldini, A., Ward, D.C., et al., Origin of Human Chromosome 2: An Ancestral Telomere-Telom-ere Fusion, Proc. Natl. Acad. Sci. USA, 1991, vol. 88, pp. 9051-9055.
Lee, C., Sasi, R., and Lin, C.C., Interstitial Localization of Telomeric DNA Sequences in the Indian Muntjac Chromosomes: Further Evidence for Tandem Chromosome Fusions in the Karyotypic Evolution of the Asian Muntjacs, Cytogenet. Cell Genet., 1993, vol. 63, pp. 156-159.
Vermeesch, J.R., De Meurichy, W., Van den Berghe, H., et al., Differences in the Distribution and Nature of the Interstitial Telomeric (TTAGGG)n Sequences in the Chromosomes of the Giraffidae, Okapi (Okapia johnstoni), and Giraffe (Giraffa camelopardalis): Evidence for Ancestral Telomeres at the Okapi Polymorphic rob (5; 26) Fusion Site, Cytogenet. Cell Genet., 1996, vol. 72, pp. 310-315.
Salvadori, S., Deiana, A., Coluccia, E., et al., Colocalization of (TTAGGG)n Telomeric Sequences and Ribosomal Genes in Atlantic Eels, Chromosome Res., 1995, vol. 3, pp. 54-58.
Liu, W.-S. and Fredga, K., Telomeric (TTAGGG)n Sequences Are Associated with Nucleolus Organizer Regions (NORs) in the Wood Lemming, Chromosome Res. 1999, vol. 7, pp. 235-240.
Pagnozzi, J.M., de Jesus Silva, M.J., and Yonenaga-Yassuda, Y., Intraspecific Variation in the Distribution of the Interstitial Telomeric (TTAGGG)n Sequences in Microureus demerarae (Marsupialia: Didelphidae), Chromosome Res., 2000, vol. 8, pp. 585-591.
Faravelli, M., Moralli, D., Bertoni, L., et al., Two Extended Arrays of a Satellite DNA Sequence at the Centromere and at the Short-Arm Telomere of Chinese Hamster Chromosome 5, Cytogenet. Cell Genet., 1998, vol. 83, pp. 281-286.
Fanning, T.G., Origin and Evolution of a Major Feline Satellite DNA, J. Mol. Biol., 1987, vol. 197, pp. 627-634.
Gruzdev, A.D., Heterochromatin and Single-Strand DNA Breaks: A Hypothesis, Genetika (Moscow), 1999, vol. 35, no. 7, pp. 869-872.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Liangouzov, I.A., Derjusheva, S.E., Saifitdinova, A.F. et al. Monomers of a Satellite DNA Sequence of Chaffinch (Fringilla coelebs L., Aves: Passeriformes) Contain Short Clusters of the TTAGGG Repeat. Russian Journal of Genetics 38, 1359–1364 (2002). https://doi.org/10.1023/A:1021679520236
Issue Date:
DOI: https://doi.org/10.1023/A:1021679520236