Abstract
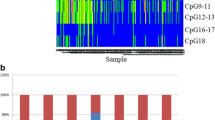
Multiplex methylation-sensitive PCR was employed in studying the methylation of CpG islands in the RB1, p16/CDKN2A, p15/CDKN2B, p14/ARF, CDH1, HIC1, and N33 5′ regions in non-small cell lung cancer (51 tumors). Methylation was observed for the two suppressor genes involved in controlling the cell cycle through the Cdk–Rb–E2F signaling pathway, RB1 (10/51, 19%) and p16 (20/51, 39%). The highest methylation frequencies were established for CDH1 (72%) and HIC1 (82%). The CpG islands of p14 and p15 proved to be nonmethylated. At least one gene was methylated in 90% (46/51) tumors and no gene, in 10% (5/51) tumors. In addition, the genes were tested for methylation in peripheral blood lymphocytes of healthy subjects. Methylation frequency significantly differed between tumors and normal cells in the case of RB1, p16, CDH1, HIC1, and N33. Gene methylation frequency was tested for association with histological type of the tumor and stage of tumor progression. Methylation index of a panel of tumor suppressor genes was established for groups of tumors varying in clinical and morphological parameters.
Similar content being viewed by others
REFERENCES
Zaletaev D.V. 2000. DNA diagnostics in oncology. Mol. Bio l 34, 671–683.
Nemtsova M.V., Zemlyakova V.V., Zhevlova A.I., et al. 2002. Methylation profile of tumor suppressor genes in various malignancies. Vestn. Inst. Mol. Med. MMA im. I.M. Sechenova. 2, 65–84.
Kim D.H., Nelson H.H., Wiencke J.K. et al. 2001. p16(INK4a) and histology-specific methylation of CpG island by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 61, 3419–3424.
Zochbauer-Muller S., Kwung M.F., Virmani A.K. et al. 2001. Aberrant promoter methylation of multiple genes in non-small cell lung cancer. Cancer Res. 61, 249–255.
Bian J., Wang Y., Kim H. et al. 1996. Suppression of in vitro tumor growth and induction of suspension cell death by tissue inhibitor of metalloproteinases-3 (TIMP-3). Carcinogenesis 17, 1805–1811.
Houle B., Rochette-Egly C., Bradley W. et al. 1993. Tumor-suppressive effect of the retinoic acid receptor β in human epidermoid lung cancer cells. Proc. Natl. Acad. Sci. USA. 90, 985–989.
Zochbauer-Muller S., Minna J., Gazdar A. 2002. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist. 7, 451–457.
Esteller M., Corn P.G., Baylin S.B. et al. 2001. A gene hypermethylation profile of human cancer. Cancer Res. 61, 3225–3229.
Zemlyakova V.V., Zhevlova, A.I., Strelnikov, V.V., et al. 2003. Abnormal methylation of several tumor suppressor genes in sporadic breast cancer. Mol. Bio l 37, 696–703.
Rathi A., Virmani A.K., Schorge J.O. et al. 2002. Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin. Cancer Res. 8, 3324–3331.
Virmani A.K., Muller C., Rathi A. et al. 2001. Aberrant methylation during cervical carcinogenesis. Clin. Cancer Res. 7, 584–589.
Virmani A.K., Tsou J.A., Siegmound K.D. et al. 2002. Hierarchical clustering of lung cancer cell lines using DNA methylation markers. Cancer Epidemiol. Biomarkers Prevention. 11, 291–297.
Lee S., Kim W.H., Jung H.Y. et al. 2002. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Amer. J. Pathol. 1, 1015–1021.
Sambrook J., Fritsh E.F., Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd end. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Strelnikov V.V., Nemtsova M.V., Chesnokova G.G. et al. 1999. Diagnostics of Martin-Bell syndrome by structural and functional changes in the 5'-untranslated region of the FMR1 gene. Mol. Bio l 33, 330–336.
Babenko O.V., Zemlyakova V.V., Saakyan S.V., et al. 2002. RB1 and CDKN2A functional defects resulting in retinoblastoma. Mol. Bio l 36, 777–783.
Baur A.S., Shaw P., Burri N. et al. 1999. Frequent methylation silencing of p15 INK4B (MTSI) and p16 INK4A (MTSI) in B-cell and T-cell lymphomas. Blood. 94, 1773–1781.
Graff J.R., Herman J.R., Myohanen S. et al. 1997. Map-ping patterns of CpG island methylation in normal and neoplastic cells implicated both upstream and down-stream region in de novo methylation. J. Biol. Chem. 227, 22322–22329.
Konishi N., Nakamura M., Kishi M. et al. 2002. DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn. J. Can. Res. 93, 767–773.
Fujii H., Biel M.A., Zhou W. et al. 1998. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 16, 2159–2164.
Li Q., Jedlicka A., Ahuja N. et al. 1998. Concordant methylation of the ER and N33 genes in glioblastoma multiform. Oncogene. 16, 3197–3202.
Weintraub S. 1996. Inactivation of tumor suppressor proteins in lung cancer. Am. J. Respir. Cell Mol. 15, 150–155.
Belinsky S.A., Nikula K.J., Palmisano W.A. et al. 1998. Aberrant methylation of p16 (INK4α) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc. Natl. Acad. Sci. USA. 95, 11891–11896.
Brabender J., Usabel H., Danenberg K.D. et al. 2001. APC gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 20, 3528–3532.
Burbee D.C., Forgacs E., Zochbauer-Muller S. et al. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancer and malignant phenotipe suppression. Natl. Cancer Inst. 93, 691–699.
Tang X., Khuri F.R., Lee J.J. et al. 2000. Hypermethylation of the death-associated protein (DAP) kinase promoter and agressiveness in stage I non-small cell lung cancer. J. Natl. Cancer Inst. 92, 1511–1516.
Palmissano W.A., Divine K.K., Saccomanno G. et al. 2000, Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 60, 5954–5958.
Zborovskaya I.B., Chizhikov V.V. 2003. Detection of molecular defects in lung cancer: The possibilities of clinical use. Novoe v terapii raka legkogo (New Findings in Therapy of Lung Cancer). Perevodchikova N.I., Ed. Moscow: Ross. Onkol. Nauchn. Tsentr im. Blokhina, 16–40.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zemlyakova, V.V., Zhevlova, A.I., Zborovskaya, I.B. et al. Methylation Profile of Several Tumor Suppressor Genes in Non-Small-Cell Lung Cancer. Molecular Biology 37, 836–840 (2003). https://doi.org/10.1023/B:MBIL.0000008351.36435.d6
Issue Date:
DOI: https://doi.org/10.1023/B:MBIL.0000008351.36435.d6