Abstract
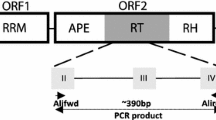
Degenerated oligonucleotide primers were used to amplify, clone, and analyze sequence heterogeneity and chromo-somal distribution of 23 PCR fragments corresponding to the reverse transcriptase domain of copia-like retrotrans-posons in rice. Of the 23 fragments 22 could be aligned by their deduced amino acid sequences and were divided into 6 groups according to the phylogenetic and Southern blot analyses. Amino acid sequence differences among the 22 aligned fragments ranged from 1 to 64%. Southern blot analysis of 10 rice accessions including indica, japonica and common wild rice, using these 23 fragments as probes, showed that copia-like retrotransposons were present in moderate to high copy numbers in all the rice genome although the exact copy number cannot be determined. The major difference revealed by southern analysis is a differentiation between the four indica varieties as one group and the four japonica varieties and the two wild rice accessions as another group. Polymorphisms were also detected among the indica and japonica varieties by major bands and repeatable minor bands. Five hybridization bands were mapped to chromosomes 3, 4, 8, and 9, respectively. All the five bands were inherited in a dominant Mendelian fashion and were not allelic with each other, indicating that the same element did not reside on the same location in different rice accessions. No transcript of the copia-like reverse transcriptase was detected on northern blot. The results suggest that the sequence heterogeneity and distributional variability of retrotransposons may be one of contributory factors causing genetic diversity in rice.
Similar content being viewed by others
References
Boeke JD, Garfinkel DJ, Styles CA, Fink GR: Ty elements transpose through an RNA intermediate. Cell 40: 491–500 (1985).
Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD: Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274 (1994).
DNASTAR Inc.: Lasergene. Biocomputing Software for the Macintosh User's Guide. A Manual for the Lasergene System, 2nd ed. (1994).
Doolittle RF, Feng D-F, Johnson MS, McClure MA: Origins and evolutionary relationships of retroviruses. Quart Rev Biol 64: 1–30 (1989).
Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A: Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucl Acids Res 20: 3639–3644 (1992).
Flavell AJ, Smith DB, Kumar A: Extreme heterogeneity of Ty1-copia group retrotransposons in plants. Mol Gen Genet 231: 233–242 (1992).
Feinberg AP, Vogelstein B: Atechnique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 (1983).
Gish W, States DJ: Identification of protein coding regions by database similarity search. Nature Genet 3: 266–272 (1993).
Grandbastien M-A, Spielmann A, Caboche M: Tntl, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337: 376–380 (1989).
Hirochika H: Activation of tobacco retrotransposons during tissue culture. EMBO J 12: 2521–2528 (1993).
Hirochika H, Fukuchi A, Kikuchi F: Retrotransposon families in rice. Mol Gen Genet 233: 209–216 (1992).
Hirochika H, Hirochika R: Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn J Genet 68: 35–46 (1993).
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M: Retrotransposons of rice involved in mutations induced by tissue culture. Proc Nalt Acad Sci USA 93: 7783–7788 (1996).
Konieczny A, Voytas DF, Cummings MP, Ausubel FM: A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127: 801–809 (1991).
Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin S-Y, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyao A, Monna L, Zhong HS, Tamura Y, Wang Z-X, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y: A300 kilobase interval geneticmap of rice including 883 expressed sequence. Nature Genet 8: 365–372 (1994).
Lin S, Min S: Rice Cultivars and Their Genealogy in China (in Chinese). Shanghai Science and Technology Press, Shanghai (1991).
Lincoln S, Daly M, Lander E: Constructing genetic maps with Mapmaker/Exp 3.0, 3rd ed. Whitehead Institute Technical Report (1992).
Mount SM, Rubin GM: Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol 5: 1630–1638 (1985).
Pearce SR, Harrison G, Li D, Heslop-Harrison JS, Kumar A, Flavell AJ: The Tyl-copia group of retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal location. Moll Gen Genet 250: 305–315 (1996).
Puissant C, Houdebine L-M: Animprovement of the single step method of RNA isolation by acid guanidinium thiocyanatephenol-chloroform extraction. Bio/technology 8: 148–149 (1990).
Saghai Maroof MA, Soliman KM, Jorgensen RA, Allard RW: Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 (1984).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Sanger F, Nicklen S, Coulson AR: DNAsequencingwith chainterminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 (1977).
Voytas DF, Cummings MP, Konieczny A, Ausubel FM, Rodermel SR: copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89: 7124–7128 (1992).
Wessler SR: Plant retrotransposons: turned on by stress. Curr Biol 6: 959–961 (1996).
Xiong Y, Eickbush TH: Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9: 3353–3362 (1990).
Zhang Q, Saghai Maroof MA, Lu TY, Shen BZ: Genetic diversity and differentiation of indica andjaponica rice detected by RFLP analysis. Theor Appl Genet 83: 495–499 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Wang, S., Zhang, Q., Maughan, P. et al. Copia-like retrotransposons in rice: sequence heterogeneity, species distribution and chromosomal locations. Plant Mol Biol 33, 1051–1058 (1997). https://doi.org/10.1023/A:1005715118851
Issue Date:
DOI: https://doi.org/10.1023/A:1005715118851