Article contents
Determination of protein–protein interactions at the single-molecule level using optical tweezers
Published online by Cambridge University Press: 10 August 2022
Abstract
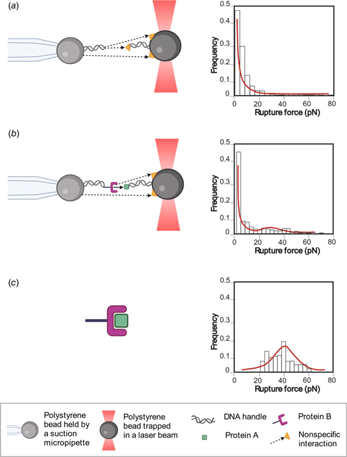
Biomolecular interactions are at the base of all physical processes within living organisms; the study of these interactions has led to the development of a plethora of different methods. Among these, single-molecule (in singulo) experiments have become relevant in recent years because these studies can give insight into mechanisms and interactions that are hidden for ensemble-based (in multiplo) methods. The focus of this review is on optical tweezer (OT) experiments, which can be used to apply and measure mechanical forces in molecular systems. OTs are based on optical trapping, where a laser is used to exert a force on a dielectric bead; and optically trap the bead at a controllable position in all three dimensions. Different experimental approaches have been developed to study protein–protein interactions using OTs, such as: (1) refolding and unfolding in trans interaction where one protein is tethered between the beads and the other protein is in the solution; (2) constant force in cis interaction where each protein is bound to a bead, and the tension is suddenly increased. The interaction may break after some time, giving information about the lifetime of the binding at that tension. And (3) force ramp in cis interaction where each protein is attached to a bead and a ramp force is applied until the interaction breaks. With these experiments, parameters such as kinetic constants (koff, kon), affinity values (KD), energy to the transition state ΔG≠, distance to the transition state Δx≠ can be obtained. These parameters characterize the energy landscape of the interaction. Some parameters such as distance to the transition state can only be obtained from force spectroscopy experiments such as those described here.
Keywords
- Type
- Review Article
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press
References
- 5
- Cited by




