Abstract
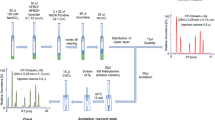
A two-dimensional LC–MS/MS system has been developed for the enantioselective determination of proline (Pro), cis-4-hydroxyproline (cis-4-Hyp) and trans-4-hydroxyproline (trans-4-Hyp) in a variety of biological samples. The amino acids were pre-column derivatized with 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F), and the NBD-derivatives were separated by a reversed-phase column (Singularity RP18) as their d plus l mixtures in the first dimension. The collected target fractions were then introduced into the second dimension where the enantiomers were separated by a Pirkle-type enantioselective column (Singularity CSP-001S) and determined by a tandem mass spectrometer (Triple Quad™ 5500). The method was validated by the standard amino acids and also by human plasma, and sufficient results were obtained for the calibration, precision and accuracy. The method was applied to human plasma and urine, bivalve tissues and fermented food/beverages. d-Pro was widely found in the human physiological fluids, bivalves and several fermented products. Although trans-4-d-Hyp was not found in all the tested samples, cis-4-d-Hyp was present in human urine and tissues of the ark shell, and further studies focusing on the origin and physiological significance of these d-enantiomers are expected.
Graphical abstract







Similar content being viewed by others
Data availability
All data are available within this published article.
References
J.J. Corrigan, Science (1969). https://doi.org/10.1126/science.164.3876.142
C. Ishii, A. Furusho, C.-L. Hsieh, K. Hamase, Chromatography (2020). https://doi.org/10.15583/jpchrom.2020.004
C. Ishii, K. Hamase, J. Pharm. Biomed. Anal. (2023). https://doi.org/10.1016/j.jpba.2023.115627
S. Karakawa, M. Harada, R. Nishimoto, Chromatography (2023). https://doi.org/10.15583/jpchrom.2023.001
T. Nishikawa, J. Chromatogr. B (2011). https://doi.org/10.1016/j.jchromb.2011.08.030
M. Katane, H. Homma, J. Chromatogr. B (2011). https://doi.org/10.1016/j.jchromb.2011.03.062
A. Furusho, K.A. Ikejiri, C. Ishii, T. Akita, M. Mita, M. Nagano, T. Ide, K. Hamase, Chromatography (2022). https://doi.org/10.15583/jpchrom.2021.020
C. Ishii, T. Akita, M. Mita, R. Konno, K. Hamase, Chromatography (2023). https://doi.org/10.15583/jpchrom.2023.002
S.H. Snyder, P.M. Kim, Neurochem. Res. (2000). https://doi.org/10.1023/a:1007586314648
A. Hashimoto, T. Nishikawa, T. Hayashi, N. Fujii, K. Harada, T. Oka, K. Takahashi, FEBS Lett. (1992). https://doi.org/10.1016/0014-5793(92)80397-y
C. Henneberger, T. Papouin, S.H.R. Oliet, D.A. Rusakov, Nature (2010). https://doi.org/10.1038/nature08673
D.S. Dunlop, A. Neidle, D. McHale, D.M. Dunlop, A. Lajtha, Biochem. Biophys. Res. Commun. (1986). https://doi.org/10.1016/s0006-291x(86)80329-1
Y. Takigawa, H. Homma, J.-A. Lee, T. Fukushima, T. Santa, T. Iwatsubo, K. Imai, Biochem. Biophys. Res. Commun. (1998). https://doi.org/10.1006/bbrc.1998.8971
Y. Nagata, H. Homma, J.-A. Lee, K. Imai, FEBS Lett. (1999). https://doi.org/10.1016/s0014-5793(99)00045-9
A. D’Aniello, M.M. Di Fiore, G.H. Fisher, A. Milone, A. Seleni, S. D’Aniello, A.F. Perna, D. Ingrosso, FASEB J. (2000). https://doi.org/10.1096/fasebj.14.5.699
R. Koga, H. Yoshida, H. Nohta, K. Hamase, Chromatography (2019). https://doi.org/10.15583/jpchrom.2019.002
R. Koga, Y. Miyoshi, Y. Sato, M. Mita, R. Konno, W. Lindner, K. Hamase, J. Chromatogr. A (2016). https://doi.org/10.1016/j.chroma.2016.07.053
R. Koga, Y. Miyoshi, E. Negishi, T. Kaneko, M. Mita, W. Lindner, K. Hamase, J. Chromatogr. A (2012). https://doi.org/10.1016/j.chroma.2012.08.075
R. Fleischmajer, L. Fishman, Nature (1965). https://doi.org/10.1038/205264a0
T. Pihlajaniemi, R. Myllylä, K.I. Kivirikko, J. Hepatol. (1991). https://doi.org/10.1016/0168-8278(91)90002-s
H.E. Jasin, C.W. Fink, W. Wise, M. Ziff, J. Clin. Invest. (1962). https://doi.org/10.1172/JCI104650
S. Adugani, G. Bannimath, P. Sastry, Biomed. Biotechnol. Res. J. (2021). https://doi.org/10.4103/bbrj.bbrj_91_21
M.A. Karsdal, S.J. Daniels, S.H. Nielsen, C. Bager, D.G.K. Rasmussen, R. Loomba, R. Surabattula, I.F. Villesen, Y. Luo, D. Shevell, N.S. Gudmann, M.J. Nielsen, J. George, R. Christian, D.J. Leeming, D. Schuppan, Liver Int. (2020). https://doi.org/10.1111/liv.14390
M. Miyanaga, T. Uchiyama, A. Motoyama, N. Ochiai, O. Ueda, M. Ogo, Skin Pharmacol. Physiol. (2021). https://doi.org/10.1159/000513988
Y. Shigemura, K. Iwai, F. Morimatsu, T. Iwamoto, T. Mori, C. Oda, T. Taira, E.Y. Park, Y. Nakamura, K. Sato, J. Agric. Food Chem. (2009). https://doi.org/10.1021/jf802785h
B. Fransson, U. Ragnarsson, Amino Acids (1999). https://doi.org/10.1007/BF01366928
Y. Huang, W. Zhang, Q. Shi, T. Toyo’oka, J.Z. Min, Food Anal. Methods (2018). https://doi.org/10.1007/s12161-018-1288-9
T. Langrock, N. García-Villar, R. Hoffmann, J. Chromatogr. B (2007). https://doi.org/10.1016/j.jchromb.2006.10.015
J. Kohama, K. Saito, H. Sakamoto, Y. Iwasaki, R. Ito, M. Horie, H. Nakazawa, Bunseki Kagaku (2007). https://doi.org/10.2116/bunsekikagaku.56.1019
S. Bernardo-Bermejo, S. Adámez-Rodríguez, E. Sánchez-López, M. Castro-Puyana, M.L. Marina, Microchem. J. (2023). https://doi.org/10.1016/j.microc.2022.108279
Y. Tojo, K. Hamase, M. Nakata, A. Morikawa, M. Mita, Y. Ashida, W. Lindner, K. Zaitsu, J. Chromatogr. B (2008). https://doi.org/10.1016/j.jchromb.2008.06.025
E. Okuma, K. Watanabe, H. Abe, Fish. Sci. (1998). https://doi.org/10.2331/fishsci.64.606
Y. Nagata, R. Masui, T. Akino, Experientia (1992). https://doi.org/10.1007/BF01919147
C. Ishii, T. Akita, M. Mita, T. Ide, K. Hamase, J. Chromatogr. A (2018). https://doi.org/10.1016/j.chroma.2018.07.076
A. Hesaka, S. Sakai, K. Hamase, T. Ikeda, R. Matsui, M. Mita, M. Horio, Y. Isaka, T. Kimura, Sci. Rep. (2019). https://doi.org/10.1038/s41598-019-41608-0
K. Okada, Y. Gogami, Y. Takeshita, T. Oikawa, Trace Nutr. Res. 29, 62–66 (2012)
Y. Miyoshi, M. Nagano, S. Ishigo, Y. Ito, K. Hashiguchi, N. Hishida, M. Mita, W. Lindner, K. Hamase, J. Chromatogr. B (2014). https://doi.org/10.1016/j.jchromb.2014.01.034
C. Ishii, T. Akita, M. Nagano, M. Mita, K. Hamase, Chromatography (2019). https://doi.org/10.15583/jpchrom.2019.011
Acknowledgements
This study was partly supported by JSPS KAKENHI Grant Numbers JP22H02752, JP22K06547 and JP23K14335. The authors appreciate KAGAMI, Inc. (Ibaraki, Osaka, Japan) for their technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ishii, C., Tojo, Y., Iwasaki, K. et al. Development of a two-dimensional LC–MS/MS system for the determination of proline and 4-hydroxyproline enantiomers in biological and food samples. ANAL. SCI. 40, 881–889 (2024). https://doi.org/10.1007/s44211-024-00530-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44211-024-00530-w