Abstract
Cancer of the lung is known to be the principal cause of tumour-linked mortality worldwide. Non-small cell lung cancer (NSCLC) is the most common and lethal type of lung cancer with close to 2.0 million reported cases and a survival rate of not less than 20% at all stages in every 5 years of diagnosis. Despite the progress made in the treatment processes, there was no significant improvement in patients with NSCLC. As such, this research aims to design (utilizing a structure-based approach) potential EGFRT790M kinase inhibitors with their DFT calculations, Drug-likeness, ADME-Toxicity properties evaluation and MD simulation. A structure-based approach was employed to design potential EGFRT790M kinase inhibitors. The DFT calculations, Drug-likeness and ADME-Toxicity properties of the newly designed potential EGFRT790M kinase inhibitors were evaluated. MD simulation of the best newly designed compound was further studied. The virtual screening performed on the investigated EGFRT790M Kinase inhibitors has identified compound 8 with the highest mole dock score of − 136.377 kcal/mol as the best hit. The structure-based strategy was adopted to design six new compounds with better affinity (between − 138.255 to − 140.552 kcal/mol) towards the EGFRT790M Kinase enzyme (3IKA). The newly designed EGFR mutant inhibitors were seen to possess better mole dock scores than AZD9291 (− 118.872 kcal/mol). Based on the predicted drug-likeness and ADMET properties, the newly designed compounds are orally bioavailable with a bioavailability score of 0.55, can be synthesized easily in the wet lab (based on their synthetic accessibility score) and possess good pharmacokinetic profiles. The MD simulation performed at 150 ns provides insight into the stability of the S2D3 compound in the binding pocket of the 3IKA target protein. The total binding free energy calculation of the studied S2D3-3IKA complex suggested that van der Waals interactions and electrostatic interactions provided the general powerful force for the binding process. The designed compound S2D3 being stable with higher affinity and best pharmacokinetic properties is therefore recommended to be synthesized in the wet lab as potential NSCLC agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is known to be the principal source of tumour-linked mortality worldwide. NSCLC and SCLC (Small cell lung cancer) are the two major classifications of cancer of the lung, respectively [1]. The ordinary and fatal kind of Lung cancer that has close to 2.10 million reported issues with an average of 22% rate of survival after diagnosis each 5 years at all stages was NSCLC [2, 3]. Close to 85% to 90% of the reported lung cases were NSCLC, and the remaining were SCLC [4]. The most widely used modalities for the mediation and treatment of the NSCLC reported were chemo, radio and targeted therapies, respectively. In spite of the progress made in the treatment processes, there was no significant improvement in patients with NSCLC [4].
In 1956, epidermal growth factor receptor (EGFR) was first uncovered by a scientist called Stanley Cohen while working in the USA at the University of Vanderbilt [5]. It is a member of the family of tyrosine kinase receptors that plays an important role in the management of cellular processes. Too much EGFR expression has been linked to many cancers, as such it was recognized as the key target for the fighting of cancer-associated ailments like NSCLC [6, 7]. Thus, designing new EGFRT790M kinase inhibitors is also among the research hotspots for the management of cancer-associated ailments most especially NSCLC [8, 9].
EGFR Tyrosine kinase inhibitors (TKIs) shared four (4) specific essential pharmacophoric features (these pharmacophoric features are: Flat hetero aromatic ring, a terminal hydrophobic head, an amino (NH-spacer) group and a hydrophobic tail, respectively) which are very important for correct binding with the active/binding site of the EGFRT790M kinase [10]. The mentioned groups are essential to occupy the adenine binding pocket, the hydrophobic region I, the space between the adenine binding region and the hydrophobic region I, and the hydrophobic region II, respectively [10]. EGFR inhibitors targeting the NSCLC, such as Erlotinib and Gefitinib, were initially designed to inhibit the growth of tumors at the EGFR ATP-binding site. Moreover, these small molecules were used as the initial choice in the management of NSCLC for the three (3) classes of EGFR mutations (the activated, classical and gatekeeper mutations) [11, 12]. Erlotinib as well as Gefitinib provide significant therapeutic aid in the management of NSCLC by protecting the EGFR tumors (in exon 19 and 21)[13]. Besides, affected persons with activating (L858R) EGFR tumors, generally recovered when treated with gefitinib and erlotinib the so-called first-generation EGFR-TKIs. Though, the period for their efficiency was actually inadequate as a result of the establishment of resistance by the gatekeeper EGFRT790M mutation that was experienced in almost half or 50% of EGFRT790M mutation patients[14]. Dacomitinib and Afatinib were subsequently established to fixed the resistance caused by the EGFRT790M mutation to the Erlotinib and Gefitinib [13, 15]. Regrettably, the efficacy of Dacomitinib and Afatinib was inadequate as a result of severe side effects which include rashes of the skin and gastrointestinal toxicity [15, 16]. In order to also fix and resolve the resistance by EGFRT790M mutation while actuality is more discriminating to the EGFRWT and experienced toxicity to the Dacomitinib and Afatinib. Osimertinib (AZD9291) and Rociletinib (CO-1686) were further designed and established. But Osimertinib and Rociletinib could not accomplish the specified role owing the immergence of resistance by the EGFR-C797S mutation [15, 17, 18].
Heterocyclic compounds as a foremost group of organic compounds are extensively used for numerous pharmacological and industrial applications. They are recognized for their biological and pharmacological futures including anti-inflammatory, antimicrobial, anticancer, antitumor, and anti-viral activities. The potential therapeutic properties of these heterocycles have encouraged medicinal chemists to synthesize a large number of novel chemotherapeutic agents [19]. Pyrimidines in general are nitrogen-containing heterocyclic organic compounds identified for anticancer, anti-HIV, antifungal, and antibacterial activities. Pyrimidines, compounds bearing this heterocycle, are common in many medicinal drugs [20]. 2,4-diaminopyrimidines derivatives are class of pyrimidines that possess diverse biological activities which include anticancer activities [21,22,23], antitumor [24], anti-bacteria activities (Bacillus anthracis) [25,26,27], anti-filarial activities [28], Anti-Tubercular Activities [29], antioxidants [30] and antidiabetics [30], anti-malaria activities [31], anti-angiogenesis activities [32]. The synthetic process to produce these compounds are reported in numerous literatures, respectively [21, 22, 32, 33]
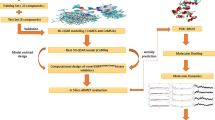
The research aims to design (utilizing a structure-based approach) potential EGFRT790M kinase inhibitors with their DFT calculations, Drug-likeness, ADME-Toxicity properties evaluation and MD simulation.
2 Methods
2.1 Sourcing of the 2,4-diaminopyrimidines derivatives
Twenty-seven (27) set of 2,4-diaminopyrimidines derivatives as NSCLC agents were sourced from the work of Cheng and co-workers [17] for the purpose of this work. The structures and names of the 27 set of 2,4-diaminopyrimidines derivatives under study were presented in Supplementary Table 1.
2.2 2D-structures sketching, optimum conformational search and compounds preparations
The 2D-conformations/structures of the obtained dataset were sketched with the help of a software established by the Cambridge University (famously known as Chemdraw) [34]. The optimum conformational search for all the 27 set of 2,4-diaminopyrimidines derivatives in this study were achieved at the MMFF with DFT at B3LYP/6-311G* theory level, respectively [15, 35].
The twenty seven (27) set of geometry optimized 2,4-diaminopyrimidines derivatives were prepared using molegro virtual docker (MVD) utilizing the default settings by choosing “if missing” to the set of parameters used by the software for compound/ligand preparation [36, 37].
2.3 EGFRT790M kinase protein and active site identification
The RCSB protein database (https://www.rcsb.org/) served as the source where the target protein (EGFRT790M kinase protein with entry code: 3IKA) in complex with WZ4002 (the co-crystalized) was retrieved from [38]. Furthermore, the active/binding sites/poses of the EGFRT790M kinase was determined/identified by visualizing the retrieved co-crystalized structure of the EGFRT790M kinase in complex with WZ4002 with discovery studio visualizer version 16.1.0.15350, respectively. The EGFRT790M kinase was further prepared before ligand-receptor docking respectively. Figure 1 depicts the 2D views of the EGFRT790M kinase in complex with WZ4002.
2.4 Molecular docking virtual screening execution and validation
The implementation of the molecular docking procedure was accomplished by picking mole dock value as the scoring function and plant value as the docking algorithm. Twenty-four Armstrong unit (24Å) was set as the size of the grid box to define the binding cavities present at the binding poses of the protein. Subsequently, default settings were preserved and maintained for all other computations in the processes [39]. The validation of docking-based virtual screening method was achieved by re-docking the best hit compound with the EGFRT790M kinase as well as computing the root means square deviation (RMSD) value of the re-docked compound in the binding pose of the EGFRT790M kinase, respectively.
2.5 Structure-based designed strategy
The strategy employed in this study is structure-based design. Structure-based design is identified as direct method of drug design which is performed when there is little or verst knowledge of the receptor target of interest. And it is simply carried out by employing either molecular docking virtual screening or fragment-based virtual screening to identify a best hit among the library of the compounds under investigation. After a successful molecular docking virtual screening, the compound noted as the hit will be reserved as template compound that will be altered/modified to come up with new compounds with better affinity towards the EGFRT790M kinase than the previous ones.
2.6 DFT computations
Density functional theory (DFT) computations at the theory level of B3LYP, using 6-31G* basis set of some of the investigated compounds were executed to have an understanding on the electronics and energy states for the some of the investigated compounds. This was achieved by calculating their lowest unoccupied molecular orbital (LUMO) and highest occupied molecular orbital (HOMO) [40,41,42]. The global reactivity descriptors (also known as quantum chemical descriptors) were also not left behind but also determined from the HOMO and LUMO energies respectively[43].
2.7 Pharmacokinetics feature evaluation
The modelling and evaluation of the pharmacokinetics (ADME-Toxic and drug-likeness) properties/features of the studied and investigated molecules were assessed using (http://structure.bioc.cam.ac.uk/pkcsm) and (http://www.swissadme.ch/index.php) web servers, respectively [34, 44, 45].
2.8 MD simulation approach
Gromacs 2019 was employed to conduct molecular dynamics (MD) simulations and estimate the binding free energies of the complex ligand-receptor [46, 47]. Parameters for both the ligand and protein were generated using the CHARMM force field. The system was built within a dodecahedron box, and a basic water model (SPC) was employed for solvation [47]. Electrical neutrality of the system was achieved by adding Na+ or Cl− counterions. Temperature coupling was maintained using the Berendsen thermostat, followed by global equilibration under limited particle number, volume, and temperature (NVT) conditions, which lasted for 20 ps at 300 K [48]. Subsequently, a constant number of particles, pressure, and temperature (NPT) was achieved by balancing the system for 1 ns. Pressure coupling at 1 atm was achieved using the Parrinello-Rahman barostat [49]. The dynamic simulation was carried out for a duration of 150 ns. The binding free energies of the compounds complexed with the target proteins were predicted using the MM-PBSA method with the gmmpbsa program [50, 51].
3 Results
3.1 Virtual screening implementation using molecular docking-based approach
The results of the virtual screening of the compounds under investigation are shown in Table 1 and Figs. 2, 3, 4, 5, 6 and 7, accordingly.
3.2 DFT computations
The result of the single-point DFT calculations is respectively shown in Table 2, and Figs. 8, 9.
3.3 Pharmacokinetics (ADME-toxic and drug-likeness) modelling and evaluation
The results of the Pharmacokinetics (ADME-Toxic and drug-likeness) modelling on the compounds under investigation are presented in Tables 3, 4, 5, 6, 7, accordingly.
3.4 Structure-based design
The results of the structure-based design on the investigated compounds are presented in Tables 8, 9, 10, Figs. 10, 11, 12, 1314 and 15 accordingly.
3.5 MD simulation
The results of the MD simulation for the best designed compounds are presented on Figs. 16, 17, 18, 19 and 20 and Table 11.
4 Discussions
4.1 Virtual screening implementation using molecular docking-based approach
The examination of the ligand-protein interactions of the twenty-seven (27) set of 2,4-diaminopyrimidines derivatives and EGFRT790M kinase with 3IKA pdb entry code was achieved via molecular docking-based approach, respectively. The EGFRT790M kinase inhibitors under investigation were ranked according their mole dock values. The mole dock values of the studied compounds were in the range of − 93.0417 to − 136.377 kcal/mol as shown in Table 2. The docking-based virtual screening executed has identified molecule 8 with a mole dock score of − 136.377 kcal/mol as the top ranked or hit among the studied compounds, then molecule 24 with a mole dock score of − 124.632 kcal/mol as the second ranked, then molecule 21 with a mole dock score of − 122.51 kcal/mol as the third ranked, then molecule 2 with a mole dock score of − 119.455 kcal/mol as the forth ranked and lastly molecule 22 with a mole dock score of − 119.436 kcal/mol as the fifth ranked also presented in Table 1.
There were four different categories of interactions observed between molecule 8, the top ranked compound among the EGFRT790M kinase inhibitors under investigation and the active site of the EGFRT790M kinase. Among the observed interactions were: non-classical carbon hydrogen bonds between the hinge residues MET793 and PRO794 amino acids with the 4-methylpiperazin-1-yl moiety of the hydrophobic head of compound 8, hydrophobic-Pi-alkyl, Pi-Pi stacked, amide-Pi stacked and alkyl interactions between LEU718, LEU844, LEU792, PHE723 and VAL726 amino acids with the central hetero aromatic system, hydrophobic head and hydrophobic tail part of molecule 8 (1H-indol-5-yl, 1H-pyrazolo[3,4-d] pyrimidin-6-yl and pyrimidine-2,4-diamine moieties), and electrostatic-Pi-anion interaction between ASP855 amino acid residue with the central hetero aromatic system (1H-pyrazolo[3,4-d] pyrimidin-6-yl moiety) of the compound, respectively (Fig. 2).
For molecule 24 in the active site of the EGFRT790M kinase (the second-best compound among the EGFRT790M kinase inhibitors under investigation), the interactions were via: 2 non-classical carbon hydrogen bonds between ASP855 amino acid and the 2 hydrogen atoms attached to the 2-dimethylamino ethyl moiety (at the hydrophobic tail), and also 1 non-classical carbon hydrogen between GLY719 amino acid and oxygen of the acrylamide group at the hydrophobic tail part of the molecule. Halogen bond interaction was also observed between GLN791 amino acid residue and the bromine on the pyrimidine ring at the central hetero aromatic system. Electrostatic (Pi-sulfur) interaction with MET790 amino acid residue was also visible between the pyrimidine ring (central hetero aromatic system) and the N-H space linker of the molecule. Furthermore, hydrophobic-Alkyl and Pi-alkyl interactions between MET790, MET793, LEU844, LEU718, VAL726, LYS745, LEU792 and ALA743 amino acid residues with the central hetero aromatic system, hydrophobic head and hydrophobic tail parts of the molecule were as well noticed as depicted by Fig. 3.
The interaction of the third-best compound 21 with the active site of EGFRT790M kinase were observed to be through classical and non-classical carbon hydrogen bonds between ASP855, LEU718 residues and the 1H-indol-6-yl (at the hydrophobic head) and the phenyl ring at the hydrophobic tail part of the molecule, hydrophobic-Pi-Sigma, Pi-Pi T-shaped, alkyl and Pi-alkyl interactions between LEU718, PHE723, ALA743, MET790, LEU844, VAL726 and LYS745 residues and 5-bromopyrimidin-2-yl (the central hetero aromatic system ) and the 1H-indol-6-yl (at the hydrophobic head part) moieties of the molecule, electrostatic-pi-cation interactions between LYS745 residue and 1H-indol-6-yl (at the hydrophobic head part) and also halogen interaction with GLN791 residue and the bromine on the pyrimidine ring (the central hetero aromatic system) of the molecule, respectively (Fig. 4).
In-line with molecule 2 (mole dock value of − 119.455 kcal/mol and re-rank value of − 100.082), among the types of interactions seen were 2 conventional carbon hydrogen bonds between 2 fluorides (F1 and F3) of the trifluoromethyl-pyrimidine moiety the central hetero aromatic system and the hinge residue MET793, 1 carbon hydrogen bonds between 1 of the fluorides (F3) of the trifluoromethyl-pyrimidine moiety the central hetero aromatic system and LEU792 residues, halogen bond between the 3 fluorides of the trifluoromethyl-pyrimidine moiety the central hetero aromatic system and GLN791 residue, electrostatic (Pi-sulfur) interaction between pyrimidine ring and MET790 residues and hydrophobic (Pi-sigma, alkyl and Pi-alkyl) interactions between the central hetero aromatic system, hydrophobic head and hydrophobic tail parts of compound 2 with ALA743, LEU844, PHE723, LEU718, LEU792, VAL726 and LYS745 acid residues, respectively (Fig. 5).
Among the types of interactions observed between compound 22 (with a mole dock score of − 119.436 kcal/mol) and the EGFRT790M kinase were both conventional and carbon hydrogen bonds between oxygen on the acrylamide moiety at the hydrophobic tail and LYS745 residue. Pi-anion and Pi-sulfur electrostatic interactions between 1-methyl-1H-indol-6-yl at the hydrophobic head part and pyrimidine ring at the central hetero aromatic system with ASP855 and MET790 residues were also observed. Furthermore, hydrophobic (Pi-Pi stacked, alkyl and Pi-alkyl) interactions between the central hetero aromatic system, hydrophobic head and hydrophobic tail parts of the compound and PHE723, LEU844, ALA743, and LEU792 acid residues were respectively noticed (Fig. 6).
Looking at the binding mode (pattern) of the reported compounds, it can be understood that all the reported compounds were seen to have similar pattern of hydrophobic interaction (that is all the central hetero aromatic system, hydrophobic head and hydrophobic tail parts participated in the hydrophobic interaction) but differ in terms of their hydrogen bonding interaction (that is only the hydrophobic head and hydrophobic tail parts of the compounds participated in the hydrogen bonding interaction with the exception of compound 2 which the central hetero aromatic system showed hydrogen bonding) and electrostatic interactions (all the compounds were seen to have formed electrostatic interaction with their central hetero aromatic system and hydrophobic head with the exception of compound 22 which formed the interaction with all the central hetero aromatic system, hydrophobic head and hydrophobic tail parts, respectively).
In order to effectively authenticate and validate the docking-based procedure, the best hit molecule 8 (co-crystallized ligand) was re-docked with the EGFRT790M kinase and 1.63Å was gotten as the root means square deviation (RMSD) value for the validation process which further validate the accuracy of the molecular docking procedure employed in this study to be reproducible and reliable. The 2D superimposed interactions of the validation process is shown in Fig. 7.
4.2 DFT computations
The DFT single point energy computations on the best hit compounds were achieved at B3LYP/6-31G* level of theory. The frontier molecular orbital energies (HOMO and LUMO energies) of a compound that postulates how important features of that compound in terms of interactions associated with transfer of charge with other species (most especially at the active site of protein/enzymes) and aids to comprehend its stability. while the energy band gap (ΔE) of a compound can be used to determine its electronic and optical properties (which demonstrates its reactivity), respectively [52, 53]. The HOMO serves as the electron-rich center (which has the potential to give electrons), while the LUMO serves as electron deficient center (which has the potential to accept electrons). The existence of the negative values in the energy gap (ΔE= LUMO-HOMO) for the best hit compounds indicate better stability that is crucial in the formation of a highly stable complex. Furthermore, the energy gap serves as a means for the identification of the most active compounds among the best hit. Moreso, a smaller energy gap between the HOMO and LUMO energies of molecules has a significant influence on their intermolecular charge transfer from HOMO to LUMO and as well their bioactivities thereby causing higher strong affinity of the molecules against their target enzyme. Whereas, a bigger energy gap between the HOMO and LUMO energies of the best hit has a negative influence on their intermolecular charge transfer from HOMO to LUMO and as well their bioactivities thereby causing weak affinity of the molecules against its target enzyme, respectively [52, 53].
Compound 8 (which is the top hit) has the lowest and smallest band/energy gap (ΔE) of 3.23 eV (Table 2), respectively. As such, it is identified as the utmost reactive compound among the reported ones (that might be the reason why it has the highest affinity toward the 3IKA-EGFR enzyme, as it possesses smaller band gap). The lower band gap is a sign of good stability for the formation of complex. Furthermore, the reactivity of a compound rises/increases with a decline/decrease in the band gap between HOMO and LUMO energies. The trend in the reactivity of the top hit in increasing order is according to: Compound 8 (3.23eV) > Compound 2 (4.16eV) > Compound 21 (4.19eV) > Compound 22 (4.21eV) > Compound 24 (4.25eV), respectively.
The HOMO and LUMO energies of the molecule were as well used to determine the quantum chemical parameters (η, δ, χ and µ) in order to further confirm the reactivity, stability and optical properties of molecule 8 (Table 2). Chemical hardness parameter (η) is associated to molecular stability and reactivity. If a molecule is hard, it means it has bigger band gap, which shows that the molecule is going to be less reactive. Whereas, in the opposite side, a soft molecule has smaller band gap, respectively [54, 55]. The frontier molecular orbital (FMO) (or HOMO and LUMO) energy diagrams of the best hit is presented in Fig. 8.
The MEP surface distribution map of compound 8 based on the polar surface areas based on quantum chemical parameters computed defines the charge circulation and dispersal for the molecule that provides well understanding of that molecule will interact with other molecules most especially it targets enzyme (protein) in a ligand-protein interactions. MEPS maps of the best hit compound 8 is depicted in Fig. 9. The colors describe the pictural behavior and magnitude of MEP surface for molecule 8. When observed from the MEPS map, red color shows negative potential zone/region and the blue color denotes the positive potential zone/region, respectively. For molecule 8, the blue region which is the positive potential zone was mostly observed/seen on their amino groups. Whereas, there were partially red colors which are the negative potential zone on the nitrogen and oxygen atoms of the compound 8.Furthermore, the greenish color which portrayed the region/zone of zero potential was dominantly distributed around the carbon atom, respectively [15, 56].
4.3 Pharmacokinetics (ADME-toxic and drug-likeness) modelling and evaluation
The web server belonging to SwissADME was used to model and evaluate the drug-likeness of the best, top ranked hit compounds identified by using the notable RO5 (MW ≤ 500, Number of HBD ≤ 5, Number of NBA ≤ 10, Calculated MLog p ≤ 5), Veber’s rule (Rotatable bond ≤10 and TPSA ≤140 Å), Egan’s rule (WLOGP ≤ 5.88 and TPSA ≤ 131.6 Å) and Muegge’s rule (200≤ MW ≤ 500, − 2 ≤ XLOGP3 ≤ 5, TPSA ≤150 Å, Rotatable bond ≤ 15 Number of HBD ≤ 5, Number of HBA ≤ 10, Calculated MLog p ≤ 5) filtering criteria [45, 57] (Tables 3, 4, 5 and 6).
The Lipinski's rule of five filters was first adopted in the filtering of the drug-likeness of the best, top ranked hit compounds identified. The evaluated drug-likeness for the best, top ranked hit compounds identified showed only one (1) failure/violation for the Lipinski's rule of five filters (MW>500) except for compound 2 which has zero failure/violation (Table 3). They were subsequently filtered using the Veber’s rule, they all have zero violation except for compound 24 which has one violation (that is it has more than 10 number of rotatable bonds) (Table 4). Table 10 presents the drug-likeness of the best, top ranked hit compounds identified following the Egan’s rule, none of the compounds was found to have violated the rule (Table 5). Furthermore, the best, top ranked hit compounds identified were further filtered utilizing the Muegge’s rule, none of the compounds have violated this filtering criteria except compound 24 which has one (1) failure/violation (that is XLOGP3 more than 5) (Table 6). Following this evaluation, the best, top ranked hit compounds identified are concluded to be drug-like compounds and orally safe by having oral bioavailability score of 0.55, respectively [58, 59].
The ADME-Toxic properties predicted for the best top ranked hit compounds identified are presented in Table 7. The human intestinal absorption (HIA) of the best, top ranked hit compounds identified was found to be within the range of 77.32 to 87.892 percent, respectively. The value of their HIA was observed to be better than the threshold accepted value of 30 percent, which clearly showed the feasibility of their absorbance in the HI. The BBB penetrability for these small molecules were observed to be < − 1 which simply mean that the hits can pass via the blood brain barrier, respectively. For their central nervous system penetration, it is > − 2 and < − 3 for the best, top ranked hit compounds except for compound 2 (− 1.951) thus, indicating that they can pass via the central nervous system, respectively. The enzyme responsible for the drug metabolism in the cell of most organisms is the cytochrome which comprises a big family of some enzymes. It is therefore, very essential to look at the metabolic pathways of these drugs in the human system (body). The most important class of cytochromes (CYP) that is saddled with the responsibility of the breaking down drugs in the body is CYP3A4. Therefore, for a drug to be good it has to inhibitor and substrate of this class of CYP3A4. For the studied compounds they were recognized as inhibitors as well as substrate of CYP3A4. This further clearly showed that they can be broken down in the human system (body). After breaking down of these compounds (drugs) in the human system (body), it is very necessary to examine how they can also be removed/excreted from the body. This brings about the evaluation of the rate at which they can be removed and their concentration in the body. They showed high value of rate of removal/excretion/total clearance but within the acknowledged threshold limit of a small molecule in the body. According to the toxicity assessment, the compounds investigated are all detected to be non-toxic with the exception of compound 22. Lastly, the overall ADME-Toxic properties of these compounds investigated showed their excellent pharmacokinetic profiles (Table 7) [60, 61].
4.4 Structure-based design of new EGFRT790M kinase inhibitors
After the DBVS (docking-based virtual screening) executed between the 27 set of 2,4-diaminopyrimidines derivatives and the EGFRT790M kinase, compound 8 with a mole dock score of − 136.377 kcal/mol was identified to be the best, top ranked hit among the investigated compounds and is therefore retained as template compound. The design strategy adopted on compound 8 was that it is divided into two main parts (R1 and R2). R1 is the part of 1-methyl-1H-indole, where hydrogen or fatty primary alcohol with long chain are introduced to replace the methyl on the nitrogen atom of the indole moiety, which is expected to show the possible action sites with the residues of the mutated EGFR kinase. While R2 is the part where sidechains of diverse dimensions were introduced to discover the utmost fit set which can have effect on the inhibitory activity of the mutated EGFR kinase. Based on this strategy, six new compounds with better affinity towards the EGFRT790M kinase were designed (Supplementary Table 2S).
4.5 Molecular docking of the newly designed EGFRT790M kinase inhibitors
Molecular docking was also performed between the newly designed EGFRT790M kinase inhibitors and the target (EGFRT790M kinase) to have insight in their ligand-protein interactions. The mole dock values of the newly designed EGFRT790M kinase inhibitors were observed within the range of − 138.255 to − 140.552 kcal/mol, as shown in Table 8. Compound S2D7 has the highest mole dock value of − 140.552 kcal/mol among the designed compounds followed by S2D3 with − 140.061 kcal/mol, then by S2D2 with a mole dock value of − 139.794 kcal/mol, then by S2D6 with a mole dock value of − 139.627 kcal/mol, then by S2D9 with a mole dock value of − 139.044 kcal/mol and by S2D10 with a mole dock value of − 138.255 kcal/mol, respectively.
The 4-methylpiperrizin-1-yl (hydrophobic tail parts) and pyrimidin-4-yl (central hetero aromatic system) moieties of compound S2D7 (with the highest affinity toward 3IKA EGFR enzyme) formed hydrogen binding force with LYS745 (1.97 Å), GLN791 (3.01 Å) and GLN791 (3.07 Å) group of amino acid residues. The central hetero aromatic system, hydrophobic head and hydrophobic tail parts of the compound interacted via hydrophobic binding interaction with VAL726 (2), PHE723, GLY857, LEU858 (2), ALA743, LEU844 & LYS875 (3) group of amino acid residues, respectively. The pyrimidin-4-yl moiety at the central hetero aromatic system of the compound formed other interaction with ASP855 amino acid at the ATP-binding site of the receptor as shown in Fig. 10.
The 1H-indol-5-yl (at the hydrophobic head parts), 1H-pyrazolo[3,4-d] pyrimidin-6-yl, 4-methylpiperazin-1-yl (at the hydrophobic tail parts) and the pyrimidine-2,4-diamine (at the central hetero aromatic system) moieties of the second-best designed compound S2D3 formed hydrogen binding force with ARG841 (2.39 Å), GLY796 (2.21 Å), ASP855 (3.09, 2.27, 2.97 Å), MET793 (2.56 Å), CYS797 (3.32 Å) group of amino acids, respectively. Then hydrophobic binding interaction with LYS745, PHE723, CYS797, ARG841, LEU718, ALA743, LEU792 (2), LEU844, VAL726 & LEU718 (2) acid residues and the central hetero aromatic system, hydrophobic head and hydrophobic tail parts of the compound was also observed. The hinge residue MET790 amino acid at the ATP-binding site of the receptor also formed other binding interaction with the phenyl ring attached to the hydrophobic tail parts of the compound as presented in Fig. 11.
The hydrogen of the amine group (N-H spacer) linking the pyrimidine ring (the central hetero aromatic system) to the indole ring (hydrophobic head), phenyl ring (at the hydrophobic tail parts) and the hydrogen of the ethyl moiety (at the hydrophobic tail parts) of the third-best compound S2D2 formed hydrogen binding force with MET793 (2.38 Å), ASP800 (2.37 Å), CYS797 (3.49 Å) amino acid groups, respectively. Hydrophobic binding interaction with these group of residues LEU799, ARG841 (2), LEU792 (2), VAL726, ALA743 (2), LYS728, LEU844 (2), LEU718 (2) and the central hetero aromatic system, hydrophobic head and hydrophobic tail parts of the compound was also observed. Other binding forces with MET790, MET793 acid residues and the indole ring of the compound were also among the noticed interactions as seen in Fig. 12.
Osimertinib (AZD9291) an approved FDA small molecule (drug) was also docked into the binding pose of the EGFRT790M kinase in order to compare the reference drug (Osimertinib/AZD9291) with the newly designed EGFRT790M kinase inhibitors. Osimertinib /AZD9291 with a mole dock value of − 118.872 kcal/mol interacted with the following amino acid residues ALA743, LEU718, LEU844, GLN791, VAL726, LYS745 and PHE723 via hydrogen bonding force, hydrophobic and electrostatic binding forces as depicted by Fig. 13. All the newly designed EGFRT790M kinase inhibitors were observed to possess better mole dock values than Osimertinib/AZD9291 (the reference drug).
4.6 DFT computations for the designed compounds
The single point energy DFT computation on the EGFRT790M kinase inhibitors newly designed was achieved at B3LYP/6-31G* level of theory to determine their stabilities and reactivities, respectively. Compound S2D6 which is among the best EGFRT790M kinase inhibitors newly designed has the lowest and smallest energy gap (ΔE) of 6.92 eV as shown on Table 8. The reactivity trend of EGFRT790M kinase inhibitors newly designed in increasing order is according to: S2D6 (6.92eV) > S2D2 (6.98eV) > S2D10 (7.28eV) > S2D19 (7.32eV) > S2D9 (7.49eV) > S2D3 (7.58eV), respectively. These claim (reactivity) of the newly designed EGFRT790M kinase inhibitors was further confirmed by computing their quantum chemical parameters (η, δ, χ and µ) values from the HOMO and LUMO energies as respectively shown on Table 8 [52, 53]. The FMO energy (HOMO and LUMO energies) diagram of the most reactive (S2D6) among the newly designed EGFRT790M kinase inhibitors are presented in Fig. 14.
The MEP surface distribution map of compound S2D6 based on the polar surface areas for the quantum chemical parameters computed defines the charge circulation and dispersal for the molecule that provides well understanding of that molecule and will interact with other molecules most especially its target enzyme (protein) in a ligand-protein interactions. MEPS maps of the compound S2D6 is depicted in Fig. 15. The colors describe the pictural behavior and magnitude of MEP surface for molecule S2D6. When observed from the MEPS map, red color shows negative potential zone/region and the blue color denotes the positive potential zone/region, respectively. On the most reactive molecule (S2D6) among the newly designed ones, the blue region which is the positive potential zone was mostly observed/seen on their amino groups. Whereas, there were partially red colors which are the negative potential zone on the nitrogen and oxygen atoms of the molecule (S2D6). Furthermore, the greenish color which portrayed the region/zone of zero potential was dominantly distributed around the carbon atom, respectively [54, 55].
4.7 ADME-toxic and drug-likeness modelling and evaluation of the newly designed compounds
The web server belonging to SwissADME was also used in modelling and evaluating the drug-likeness of the newly designed EGFRT790M kinase inhibitors by adopting the Lipinski's rule of five, Veber’s rule, Egan’s rule and Muegge’s rule filtering criteria (Table 9) [45]. The drug-likeness of these newly designed EGFRT790M kinase inhibitors were initially screened adopting the Lipinski's filters, the modelled and evaluated drug-likeness of the newly designed EGFRT790M kinase inhibitors showed only one (1) failure for all the compounds. They were subsequently filtered using the Veber’s rule, they all have one violation except for compound S2D3, S2D7 with zero violation and S2D6 with 2 violations. Furthermore, the drug-like features of the designed compounds following the Egan’s rule has shown that all have one violation with the exception of S2D3 which has zero violation. Moreover, the Muegge’s rule showed one violation for S2D2 and S2D10, two violations for S2D6 and zero violation for S2D3, S2D7 and S2D9, respectively. According to the used drug-like filtering criteria, the newly designed EGFRT790M kinase inhibitors were established to be drug-like compounds and orally bioavailable with bioavailability score of 0.55. Furthermore, based on the values of the synthetic accessibility they can all be synthesized easily in the wet lab [58, 59].
The ADMET features predicted for the designed molecules are presented in Table 10. The HIA of the EGFRT790M kinase inhibitors newly designed were all observed within the range of 63.5 to 78.678 %, respectively. Their HIA values were found to be greater than the threshold accepted value which is 30% set for the assessment of this feature. The blood brain barrier penetration of these newly designed EGFRT790M kinase inhibitors were detected to be < − 2 except for S2D3 which was < − 1 simply mean they can poorly pass via the blood brain barrier with the exception of S2D3, respectively. Their central nervous system penetration value was > − 3. And for this reason, they can poorly pass through the central nervous system with the exception of S2D3 which was found to be >− 2 but < − 3 (− 2.774). This clearly showed that it can better pass through the central nervous system, respectively. The enzyme responsible for the drug metabolism in the cell of most organisms is the cytochrome which comprises a big family of some enzymes. It is therefore, very essential to look at the metabolic pathways of these drugs in the body. The most important class of cytochromes (CYP) that is saddled with the responsibility of the breaking down drugs in the body is CYP3A4. Therefore, for a drug to be good it has to inhibitor and substrate of this class of CYP3A4. For the studied compounds they were recognized as inhibitors as well as substrate of CYP3A4. This further clearly showed that they can be broken down in the body. After breaking down of these EGFRT790M kinase inhibitors newly designed (drugs) in the human system (body), therefore, it is very essential to examine how they can also be removed/excreted from the body. This brings about the evaluation of the rate at which they can be removed and their concentration in the body. They all displayed average value of excretion/total clearance but within the acknowledged threshold limit of a small molecule in the body. These EGFRT790M kinase inhibitors newly designed are noticed to be non-toxic as ascertained by the toxicity test performed. In conclusion, the overall ADME-Toxic properties of these EGFRT790M kinase inhibitors newly designed portrayed their good pharmacokinetic profiles most especially S2D3 which was found to be better than others [60, 61].
4.8 MD simulation
To support the results of the molecular docking performed, the best design compound (S2D3) with the higher affinity towards the protein (3IKA) was subjected to molecular dynamics simulations to clarify the dynamic behavior of the interactions involved between the ligand-receptor complex for 150 ns.
The root means square deviation (RMSD) serves as a measurable/quantitative future/parameter used to evaluate and interpret the stability of the movement of the receptor’s atoms chain when the ligand is present. The RMSD plots for the S2D3-3IKA protein complex are presented in Fig. 16.
The RSMD fluctuations of the S2D3-3IKA protein complex were in a range between 0.13 and 0.38Å. It is observed from the plot that the S2D3 ligand shows stability during the simulation trajectory after 20 ns around the least weak RMSD value of approximately 0.3Å, indicating that the compound S2D3 is very stable in the 3IKA protein binding site, respectively.
Figure 17 depicts the root means square fluctuation (RMSF) analysis of the S2D3-3IKA target protein complex to evaluate the protein stability and comprehend the flexibility of the 3IKA target receptor, the average fluctuation per 3IKA receptor backbone residue. It is observed from the figure that there was less conformational flexibility for the S2D3 bound system. The residue fluctuations of the 3IKA target protein are very low (around 0.05Å) in the S2D3 bound system suggesting its stability in the interactions formed.
The radius of gyration (Rg) shows the capability and form of S2D3-3IKA target protein complex structures throughout the simulation time. It is an additional indicator of ligand stability as shown by the Rg plot. The S2D3-3IKA target protein complex showed Rg of 21.34 Å at the beginning of the first 20 ns before it stabilizes at the rest of the time at Rg of 21.36Å, respectively. The radius of the gyration plot is presented in Fig. 18.
The hydrogen bonding was further estimated in the 3IKA target protein in the presence of the S2D3 ligand investigated. The maximum number of H bonds observed throughout the simulation time was found to be three (3) for the 3IKA target protein as depicted in Fig. 19.
Solvent-accessible surface (SASA) was further investigated to measure fluctuations in hydrophilic and hydrophobic residues in the 3IKA target protein. Remarkably, the 3IKA target protein displayed good stability with the S2D3 ligand. The 3IKA target protein in the presence of an S2D3 ligand disclosed stable SASA values between 1.70 to 1.76Å2 throughout the simulation time as depicted in Fig. 20, respectively.
The binding free energy of the S2D3-3IKA target protein complex was predicted by the utilization of the molecular mechanics Poisson-Boltzmann surface analysis (MMPBSA) method. The MMPBSA investigation was examined over the entire 150 ns trajectory. The total binding energies of the S2D3-3IKA target protein complex are presented in Table 11. As presented in Table 11, S2D3 was observed to have a negative binding free energy value of − 26.627 kJ/mol. Moreover, van der Waals and electrostatic interactions and SASA (nonpolar solvation) energy offered a negative contribution to the total free binding energy, whereas polar solvation energy displayed a positive contribution to the total free binding energy. In relation to the negative contribution, the van der Waals energy contributed much more than the electrostatic actions and the SASA energies for the 3IKA target protein, this result shows that the hydrophobic interaction played a vital role in the stability of the S2D3 ligand in the 3IKA target protein. Normally, the prediction of the binding free energy using the MM/PBSA method showed that the SASA (nonpolar solvation) energy, van der Waals and electrostatic energies contributed to the stability of the simulated S2D3-3IKA target protein complex [62,63,64].
5 Conclusion
The molecular docking-based virtual screening carried out has identified compound 8 with the highest mole dock scores of − 136.377 kcal/mol as the top ranked hit compound and retained as template scaffold to design new EGFRT790M kinase. Six new EGFRT790M kinase inhibitors were designed utilizing structure-based design strategy with mole dock values within the range of − 138.255 to − 140.552 kcal/mol, respectively. All the newly designed EGFRT790M kinase inhibitors were then compared with Osimertinib/AZD9291 (the reference drug) with a mole dock value of − 118.872 kcal/mol and found to have better mole dock values. The DFT computations performed identified compound S2D6 as the most reactive compound among the designed compounds with the smallest energy gap (ΔE) of − 6.92 eV. The modelled drug-likeness and ADME-Toxic properties of the newly designed EGFRT790M kinase inhibitors showed their good pharmacokinetic profiles with bioavailability score of 0.55 and synthetic accessibility score between the range of 4.02 to 4.99, respectively. Compound S2D3 was further subjected to MD simulation and binding free energy calculation by MM-GBSA method and justifies the stability of the S2D3-3IKA target protein complex system and confirms the molecular docking result. Compound S2D3 is therefore recommended to be synthesize in the wet lab as potential non-small cell lung cancer therapeutic drug.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Bistrović A et al (2018) Design, synthesis and biological evaluation of novel benzimidazole amidines as potent multi-target inhibitors for the treatment of non-small cell lung cancer. Eur J Med Chem 143:1616–1634
Khuri F (2016) Lung cancer and other pulmonary neoplasms. Goldman-Cecil Med 2:1303–1313
Tièche CC et al (2016) Prolonged pemetrexed pretreatment augments persistence of cisplatin-induced DNA damage and eliminates resistant lung cancer stem-like cells associated with EMT. BMC Cancer 16(1):1–15
Wang D et al (2022) A multifunctional nanotheranostic agent potentiates erlotinib to EGFR wild-type non-small cell lung cancer. Bioact Mater 13:312–323
Sato JD et al (2018) A tribute to Dr. Gordon Hisashi Sato (December 24, 1927–March 31, 2017). In Vitro Cell Dev Biol-Animal 54(3):177–193
Hanan EJ et al (2016) 4-Aminoindazolyl-dihydrofuro [3, 4-d] pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase. Bioorgan Med Chem Lett 26(2):534–539
Ibrahim MT, Uzairu A, Uba S, Shallangwa GA (2021) Design of more potent quinazoline derivatives as EGFRWT inhibitors for the treatment of NSCLC: a computational approach. Future J Pharm Sci 7(1):1–11
Othman IM et al (2021) New pyrimidine and pyrazole-based compounds as potential EGFR inhibitors: Synthesis, anticancer, antimicrobial evaluation and computational studies. Bioorgan Chem 114:105078
Tu Y et al (2017) Design, synthesis, and docking studies of quinazoline analogues bearing aryl semicarbazone scaffolds as potent EGFR inhibitors. Bioorgan Med Chem 25(12):3148–3157
Sobh EA et al (2023) Design, synthesis, docking, MD simulations, and anti-proliferative evaluation of thieno [2, 3-d] pyrimidine derivatives as new EGFR inhibitors. J Enzym Inhib Med Chem 38(1):2220579
Passaro A et al (2021) Recent advances on the role of EGFR tyrosine kinase inhibitors in the management of NSCLC with uncommon, non exon 20 insertions, EGFR mutations. J Thorac Oncol 16(5):764–773
Hsu W-H, Yang J-H, Mok T, Loong H (2018) Overview of current systemic management of EGFR-mutant NSCLC. Ann Oncol 29:i3–i9
Song Z et al (2017) Synthesis and biological evaluation of morpholine-substituted diphenylpyrimidine derivatives (Mor-DPPYs) as potent EGFR T790M inhibitors with improved activity toward the gefitinib-resistant non-small cell lung cancers (NSCLC). Eur J Med Chem 133:329–339
Ibrahim MT, Uzairu A, Shallangwa GA, Uba S (2020) Lead Identification of Some Anti-Cancer Agents with Prominent Activity Against Non-small Cell Lung Cancer (NSCLC) and Structure-Based Design. Chem Afr 3(4):1023–1044
Pawara R, Ahmad I, Surana S, Patel H (2021) Computational identification of 2, 4-disubstituted amino-pyrimidines as L858R/T790M-EGFR double mutant inhibitors using pharmacophore mapping, molecular docking, binding free energy calculation, DFT study and molecular dynamic simulation. In Silico Pharmacol 9(1):1–22
Patel H, Pawara R, Ansari A, Surana S (2017) Recent updates on third generation EGFR inhibitors and emergence of fourth generation EGFR inhibitors to combat C797S resistance. Eur J Med Chem 142:32–47
Chen L et al (2017) Structure-based design and synthesis of 2, 4-diaminopyrimidines as EGFR L858R/T790M selective inhibitors for NSCLC. Eur J Med Chem 140:510–527
Karnik KS et al (2021) Computational and Synthetic approach with Biological Evaluation of Substituted Quinoline derivatives as small molecule L858R/T790M/C797S triple mutant EGFR inhibitors targeting resistance in Non-Small Cell Lung Cancer (NSCLC). Bioorgan Chem 107:104612
Zarenezhad E, Farjam M, Iraji A (2021) Synthesis and biological activity of pyrimidines-containing hybrids: Focusing on pharmacological application. J Mol Struct 1230:129833
Basha J, Goudgaon NM (2021) A comprehensive review on pyrimidine analogs-versatile scaffold with medicinal and biological potential. J Mol Struct 1246:131168
Ma W-F et al (2014) One-pot synthesis and antiproliferative activity of novel 2, 4-diaminopyrimidine derivatives bearing piperidine and piperazine moieties. Eur J Med Chem 84:127–134
Sander K et al (2009) 2, 4-Diaminopyrimidines as histamine H4 receptor ligands—Scaffold optimization and pharmacological characterization. Bioorgan Med Chem 17(20):7186–7196
Mahapatra A, Prasad T, Sharma T (2021) Pyrimidine: A review on anticancer activity with key emphasis on SAR. Future J Pharm Sci 7(1):1–38
Hu G et al (2019) Design, synthesis and biological evaluation of novel 2, 4-diaminopyrimidine derivatives as potent antitumor agents. New J Chem 43(25):10190–10202
Nammalwar B et al (2015) Modified 2, 4-diaminopyrimidine-based dihydrofolate reductase inhibitors as potential drug scaffolds against Bacillus anthracis. Bioorgan Med Chem 23(1):203–211
Ajibade PA, Idemudia OG (2013) Synthesis, characterization, and antibacterial studies of Pd (II) and Pt (II) complexes of some diaminopyrimidine derivatives. Bioinorgan Chem Appl 2013:1–8
Songsungthong W et al (2021) A novel bicyclic 2, 4-diaminopyrimidine inhibitor of Streptococcus suis dihydrofolate reductase. PeerJ 9:e10743
Sharma R et al (2013) Exploration of 2, 4-diaminopyrimidine and 2, 4-diamino-s-triazine derivatives as potential antifilarial agents. Parasitology 140(8):959–965
Ouyang Y et al (2017) Synthesis of 2, 4-diaminopyrimidine core-based derivatives and biological evaluation of their anti-tubercular activities. Molecules 22(10):1592
Mahnashi MH et al (2022) Phytochemistry, anti-diabetic and antioxidant potentials of Allium consanguineum Kunth. BMC Complement Med Ther 22(1):154
Hoarau M et al (2021) Discovery of new non-pyrimidine scaffolds as Plasmodium falciparum DHFR inhibitors by fragment-based screening. J Enzym Inhib Med Chem 36(1):198–206
Wang S et al (2021) Design, synthesis, and biological evaluation of 2, 4-diamino pyrimidine derivatives as potent FAK inhibitors with anti-cancer and anti-angiogenesis activities. Eur J Med Chem 222:113573
Li J et al (2021) Design, synthesis and biological evaluation of novel 2, 4-diaryl pyrimidine derivatives as selective EGFRL858R/T790M inhibitors. Eur J Med Chem 212:113019
Mills N (2006) ChemDraw Ultra 10.0 CambridgeSoft, 100 CambridgePark Drive, Cambridge, MA 02140. www. cambridgesoft. com. Commercial Price: 1910fordownload, 2150 for CD-ROM; Academic Price: 710fordownload, 800 for CD-ROM. J Am Chem Soc 128:13649
Kohn W, Becke AD, Parr RG (1996) Density functional theory of electronic structure. J Phys Chem 100(31):12974–12980
Ghamali M et al (2016) The inhibitory activity of aldose reductase of flavonoid compounds: Combining DFT and QSAR calculations. J Taibah Univ Sci 10(4):534–542
Ibrahim MT, Uzairu A, Shallangwa GA, Uba S (2020) Structure-based design and activity modeling of novel epidermal growth factor receptor kinase inhibitors; an in silico approach. Sci Afr 9:e00503
Chang J et al (2017) Development of a series of novel 4-anlinoquinazoline derivatives possessing quinazoline skeleton: Design, synthesis, EGFR kinase inhibitory efficacy, and evaluation of anticancer activities in vitro. Eur J Med Chem 138:669–688
Abdullahi M et al (2020) Virtual molecular docking study of some novel carboxamide series as new anti-tubercular agents. Eur J Chem 11(1):30–36
Panwar U, Singh SK (2021) Atom-based 3D-QSAR, molecular docking, DFT, and simulation studies of acylhydrazone, hydrazine, and diazene derivatives as IN-LEDGF/p75 inhibitors. Struct Chem 32(1):337–352
Chinnasamy S et al (2020) Molecular docking and molecular dynamics simulation studies to identify potent AURKA inhibitors: assessing the performance of density functional theory, MM-GBSA and mass action kinetics calculations. J Biomol Struct Dyn 38(14):4325–4335
Abdelsalam EA et al (2019) Synthesis and in vitro anticancer evaluation of some fused indazoles, quinazolines and quinolines as potential EGFR inhibitors. Bioorgan Chem 89:102985
Omoboyowa DA (2022) Virtual Screening of Phyto-compounds from Blighia sapida as Protein Tyrosine Phosphatase 1B Inhibitor: A Computational Approach Against Diabetes. Chem Afr 5:1–11
Hadni H, Elhallaoui M (2020) 3D-QSAR, docking and ADMET properties of aurone analogues as antimalarial agents. Heliyon 6(4):e03580
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
El Mchichi L et al (2022) 3D-QSAR study, docking molecular and simulation dynamic on series of benzimidazole derivatives as anti-cancer agents. J Indian Chem Soc 99(9):100582
Tabti K, El Mchichi L, Moukhliss Y, Singh G, Sbai A (2022) CoMFA Topomer, CoMFA, CoMSIA, HQSAR, docking molecular, dynamique study and ADMET study on phenyloxylpropyl isoxazole derivatives for coxsackie virus B3 virus inhibitors activity. Moroccan J Chem 10(4):703–725
Hajji H et al (2021) In silico investigation on the beneficial effects of medicinal plants on diabetes and obesity: molecular docking, molecular dynamic simulations, and ADMET studies. Biointerface Res Appl Chem 11(5):6933–6949
Saito H, Nagao H, Nishikawa K, Kinugawa K (2003) Molecular collective dynamics in solid para-hydrogen and ortho-deuterium: The Parrinello–Rahman-type path integral centroid molecular dynamics approach. J Chem Phys 119(2):953–963
Tabti K et al (2022) Molecular modelling of antiproliferative inhibitors based on SMILES descriptors using Monte-Carlo method, docking, MD simulations and ADME/Tox studies. Mol Simul 48(17):1575–1591
Kumari, R., Kumar, R., Consortium, O.S.D.D., Lynn, A (2014) g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inform Model 54(7):1951–1962
Abdullahi M et al (2023) Structure-based drug design, molecular dynamics simulation, ADMET, and quantum chemical studies of some thiazolinones targeting influenza neuraminidase. J Biomol Struct Dyn 1:1–15
Chaudhari B, Patel H, Thakar S, Ahmad I, Bansode D (2022) Optimizing the Sunitinib for cardio-toxicity and thyro-toxicity by scaffold hopping approach. Silico Pharmacol 10(1):10
Abdizadeh R, Hadizadeh F, Abdizadeh T (2023) Evaluation of apigenin-based biflavonoid derivatives as potential therapeutic agents against viral protease (3CLpro) of SARS-CoV-2 via molecular docking, molecular dynamics and quantum mechanics studies. J Biomole Struct Dyn 41(13):5915–5945
Abisha W, Dhas DA, Balachandran S, Joe IH (2023) Molecular structure, spectroscopic elucidation (FT-IR, FT-Raman, UV-Visible and NMR) with NBO, ELF, LOL, RDG, Fukui, drug likeness and molecular docking analysis on dimethomorph. Polycycl Aromat Compound 43(5):3988–4031
Omoboyowa DA, Singh G, Fatoki JO, Oyeneyin OE (2022) Computational investigation of phytochemicals from Abrus precatorius seeds as modulators of peroxisome proliferator-activated receptor gamma (PPARγ). J Biomole Struct Dyn 1:1–15
Ahmed A et al (2022) Novel adamantyl clubbed iminothiazolidinones as promising elastase inhibitors: design, synthesis, molecular docking ADMET and DFT studies. RSC Adv 12(19):11974–11991
Abuelizz HA et al (2021) Biological Evaluation of 4-(1H-triazol-1-yl) benzoic Acid Hybrids as Antioxidant Agents: In Vitro Screening and DFT Study. Appl Sci 11(24):11642
Demir Y, Ceylan H, Türkeş C, Beydemir Ş (2022) Molecular docking and inhibition studies of vulpinic, carnosic and usnic acids on polyol pathway enzymes. J Biomol Struct Dyn 40(22):12008–12021
Kraithong S, Teerapattarakan N, Balasubramanian B, Issara U (2022) Bioactive compounds in tea: Effect of imbalanced intake on digestive enzymes activity, cytochrome inhibition and drug interaction. South Afr J Bot 150:58–68
Li H, Sheng Y, Li W, Yuan L (2022) Recent advances in molecular fluorescent probes for CYP450 sensing and imaging. Chemosensors 10(8):304
Tabti K et al (2023) Molecular modeling study of pyrrolidine derivatives as novel myeloid cell leukemia-1 inhibitors through combined 3D-QSAR, molecular docking, ADME/Tox and MD simulation techniques. J Biomol Struct Dyn 1:1–17
Baammi S, Daoud R, El Allali A (2023) Assessing the effect of a series of mutations on the dynamic behavior of phosphite dehydrogenase using molecular docking, molecular dynamics and quantum mechanics/molecular mechanics simulations. J Biomol Struct Dyn 41(9):4154–4166
Tabti K et al (2023) Design and development of novel spiro-oxindoles as potent antiproliferative agents using quantitative structure activity based Monte Carlo method, docking molecular, molecular dynamics, free energy calculations, and pharmacokinetics/toxicity studies. J Mol Struct 1284:135404
Acknowledgements
The authors sincerely acknowledged Ahmadu Bello University, Zaria for its technical support in the course of this research.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
MTI and SA formulated, conceptualized, and designed the research. MTI did the program using software, organized the data, and analyzed the result. MTI wrote the final drafts of the paper. KT performed MD simulation. AST, AM and AMD reviewed and edited the language of the draft. All authors accepted the final draft and are responsible for the manuscript’s content and similarity index.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ibrahim, M.T., Tabti, K., Abdulsalam, S. et al. Discovery of new 2,4-diaminopyrimidines derivatives as EGFRT790M kinase inhibitors: a structure-based approach with DFT calculation, drug-likeness, ADME-toxicity properties evaluation and MD simulation. J.Umm Al-Qura Univ. Appll. Sci. (2024). https://doi.org/10.1007/s43994-023-00099-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43994-023-00099-6