Abstract
Biofilm contributes hugely to the persistence of typhoid fever in human population and quorum sensing (QS) is an integral mechanism involved in biofilms. Interruption of the QS network has therefore been put forward as one of the important anti-virulence strategies. Methanol extract of Psidium guajava leaves has been confirmed to possess antibacterial and anti-biofilm activities against Salmonella Typhi. This study therefore aimed at investigating the interactions of phytocompounds previously identified in the extract with selected QS proteins of S. Typhi in silico. Appropriate formats of compounds were retrieved and translated using online web servers. Quantitative estimate of drug-likeness, as well as absorption, distribution, metabolism, excretion and toxicity profiles of the compounds, were assessed on ADMETlab 2.0. Three-dimensional structures of two QS proteins of S. Typhi were obtained from Protein Data Bank while others were modelled on SWISS-MODEL. Selected compounds (ligands) were docked with the four proteins via AutoDock 1.5.6 and analyzed on Discovery studio. Eight, out of the seventy-two, phyto-compounds of methanol extract of P. guajava possess desirable drug-likeness (QED > 0.67). Three of them have toxic characteristics and thus, were removed from further consideration. Molecular docking revealed that, of the 5 ligands docked against the proteins, only Benzeneethanamine, 4-methoxy- and Cyclopentadecanone, 2-hydroxy- had affinities for the proteins of interest. The affinity of Cyclopenftadecanone,2-hydroxy- for each of the proteins is higher than that of Benzeneethanamine,4-methoxy- with hydrogen bonds contributing significantly to the interactions. Benzeneethanamine, 4-methoxy- and Cyclopentadecanone,2-hydroxy- from Psidium guajava leaves possess inhibitory properties against QS proteins of S. Typhi.
Similar content being viewed by others
1 Introduction
Salmonella enterica serovar Typhi (S. Typhi) is a foodborne pathogen responsible for causing typhoid fever, a disease that constitutes a major global health concern [3]. A major feature of S. Typhi infection is its host restriction to humans which means that the infection cycle begins and ends within the human host [16]. Typhoid fever has persisted in the human population through the development of carrier status by some infected individuals who continue to transmit the causative bacterium even after recovering from the clinical symptoms of the disease. Formation of biofilm in the epithelial cells of the gall bladder as well as gallstones, where present, have been implicated in the carrier status and antimicrobial resistance in typhoid fever [18, 19].
A biofilm is a community (colony) of microorganisms that adhere to each other and to a solid (inert or live) surface that is encased in a self-initiated protective extracellular matrix, containing polysaccharides, proteins, extracellular microbial DNA and enzymes, ions and nutrients from the environment [4, 7, 9, 19]. Biofilm contributes to the failure of anti-infective therapy by serving as a reservoir of resistant microorganisms as bacteria in biofilms are up to 103 more resistant to antibiotics than their planktonic counterparts [36]. Due to the heterogeneous nature of biofilms, several mechanisms have been reported for increased antimicrobial resistance in biofilm structures. These include (1) Low diffusion of antibiotics across the extracellular matrix resulting in low intracellular concentration of antibiotic(s). This is due to the presence of glycocalyx, an integral part of bacterial biofilm that accumulates antimicrobial substances and limits the transportation to the organisms within the colony [38], (2) phenotypic and physiological changes due to slow growth rate and starvation responses (oxygen, nutrient deprivation or environmental stress- antibiotics are not active against dormant cells; (3) the expression of efflux pumps (EP) that decrease intracellular antimicrobial concentration; (4) the emergence of persister cells, a biofilm-specific phenotype which are multi drug-tolerant cells that have not acquired genetic resistance [4, 7, 13, 14], Soto et al. [39]; (5) alteration in gene expression and (6) increase in horizontal transmission of resistance elements (Koopman et al., [25]). S. enterica is known to form biofilms on both living and non-living surfaces [19]. This characteristic has been reported to be very important for the survival of pathogenic bacteria within the human body as both innate and adaptive immune responses might not be able to eliminate pathogens within the well-established biofilm [9]. Enteric pathogens, salmonella inclusive, are resistant to the bile's detergent-like property and therefore can survive in the gall bladder. In addition, they respond to the bile environment by regulating other resistance-related genes [19]. An important mechanism for the formation of bacterial biofilm is quorum sensing [37].
Quorum sensing or signalling (QS) is a cell population density-dependent phenomenon that defines intra- and inter-specie cellular communications between bacteria which enables them to respond and adjust to their environment by regulation of certain gene expressions [31,32, 37]. In Salmonella spp, QS is responsible for bile adaptation, enhancement of persister cells and biofilm (Walawalkar et al., [43]), expression of flagella genes [11] and normal expression of salmonella pathogenicity island-1 (SPI-1), a virulence factor), [10]. QS is also involved in the production of secondary metabolites and stress adaptation mechanisms such as bacterial competition systems including the secretion systems (SS) [32]. QS in bacteria is mediated by the production of signalling protein molecules referred to as auto-inducers (AI). AIs are normally produced at an elemental level during the exponential growth phase of bacteria. However, at a cell density-dependent threshold concentration (i.e. the quorum level), they accumulate intra-cellularly via ATP-binding cassette (ABC) transporter and activate receptor proteins (transcription regulators) leading to a cascade of processes that results in the regulation of certain gene expression [46].
The QS system of S. Typhi comprises LuxS/AI-2/luxR homolog SdiA. Salmonella spp produces AI-2 during the exponential phase of growth in a reaction catalyzed by AI-2 synthase (LuxS protein). LuxS catalyzes the final step in the biosynthesis of AI-2 from S-adenosyl methionine yielding 4,5-dihydroxy-2,3-pentanedione (DPD). AI-2 is then obtained from (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (Walawalkar et al., [43]). Accumulation of AI-2 occasioned by a high-density cell population up-regulates the expression of an ATP-binding cassette (ABC) transporter (comprising of LsrA, LsrB, LsrC and LsrD), which transports AI-2 back into the cell [42]. In the cell, AI-2 is phosphorylated by a kinase (LsrK), and further modified by LsrF and LsrG (Vendeville et al., [41]). The resulting molecule binds transcription factors (SdiA) and regulates several genes pertaining to the formation of biofilm and other virulence factors of the bacterium.
Due to the integral involvement of QS in the formation and maintenance of biofilm, interruption of the QS network has been put forward as one of the important anti-virulence strategies [36] used to target biofilm-forming bacterial by any of the following means: (1.) inhibition of synthesis of QS signalling molecules,(2.) blocking the transport of AI into the cell; (3.) inhibition of intracellular processing of AI and (4.) inhibition of receptor proteins.
There abounds, in the literature, evidence of anti-bacterial and anti-biofilm properties of plant materials that can be harnessed for combating antimicrobial resistance of pathogenic organisms by interrupting the biofilm formation system. For instance, various parts of P. guajava have been reported for anti-biofilm activities against Staphylococcus aureus (250 μg/mL of pulp extract) [12], Streptococcus mutans and Streptococcus gordonii (0.78 mg/mL of leave extract) [28]. Fu et al. also reported that aqueous extract of Herba patriniae exhibited an inhibitory effect on almost all the studied biofilm-related genes of Pseudomonas aeruginosa with a significant reduction in the formation of biofilm as well as an alteration in the structure of the mature biofilms. Exopolysaccharide production and swarming motility were also impaired in the disfavour of biofilm formation [15].
Psidium guajava L (family- Myrtaceae; common name- guava), a fruit-bearing plant of the tropics, has been variously reported for traditional treatment of typhoid fever [30] as well as other antibacterial activities. Methanol extract of P. guajava leaves has also been confirmed to possess antibacterial and anti-biofilm activities against S. Typhi [1]. Previously, we identified the phyto-compounds in the methanol extract of P. guajava leaves and their interactions with a protein involved in replication of S. Typhi [2]. In this study, we investigate the interaction between those compounds and key proteins involved in QS in the typhoid bacteria to identify molecules that could be investigated further in the quest for a new drug for tackling typhoid fever.
2 Methods
An overview of the stages involved in this study is presented in Fig. 1. The final docking scores of these phytochemicals are compared to those of ciprofloxacin which is currently used for the treatment of typhoid fever.
2.1 Ligands preparation
The ligands are compounds previously identified in the methanol extract of P. guajava leaves [2]. All seventy-two identified compounds were considered and coded 1–72. The Simplified Molecular-Input Line-Entry Specification System (SMILES) formats of the ligands were obtained from PubChem [22] and translated to PDB format using the CACTUS online translator [8]. Ligands were converted to PDBQT formats on OpenbabelUI software.
2.2 Drug-likeness and ADMET profiling of ligands
The drug-likeness or otherwise of each compound was assessed via the QED (Quantitative Estimate of Drug-likeness). ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) profiles of compounds with desirable QED values were also predicted. Both QED and ADMET were evaluated using ADMETlab 2.0 online webserver [47]. Only compounds with desirable QED and no predicted toxicity were selected for molecular docking.
2.3 Preparation of proteins
2.3.1 Retrieval from Protein Data Bank
The three-dimensional crystal structures (Fig. 2) of two QS proteins of S. Typhi were obtained from PDB (Protein Data Bank) (LuxS, PDB Id: 5v2w; LsrB, PDB Id: 5GTA).
2.4 Homology modelling
Two proteins (LsrK and SdiA) whose 3-D structures were not available in PDB at the time of retrieval, were modelled using the SWISS-MODEL homology webserver [44]. Amino acid FASTA sequences of LsrK (530aa, Accession: QKX96401.1) and SdiA (240aa, Accession: QKX97843.1) were retrieved from the website of the National Centre for Biotechnology Information for modelling. SWISS-MODEL performs all the stages of modelling viz: (i.) template identification and selection,(ii.) alignment target-template sequence; (iii.) model building and refinement and (iv.) model evaluation [34].
2.5 Molecular docking
To study the interaction between selected compounds (ligands) and the selected QS proteins of S. Typhi (protein), molecular docking was carried out using AutoDock Vina 1.5.6 software. The proteins were first prepared by adding polar hydrogen atoms, calculating Kollman charges (Table 1) and saving them in PDBQT format on the AutoDock tool [40]. The grid box was set to cover the entire surface of the protein to allow for blind docking. The prepared and optimized ligands were blindly docked in the grid box of the protein to allow ligands to find any favourable binding site on the protein. Ligands were ranked based on their binding affinities with higher negative values signifying better affinity. Ligand–protein interactions were visualized using Discovery Studio Visualizer 2020 software.
3 Results
3.1 QED of compounds
The QED of compounds, calculated based on eight physicochemical properties (molecular weight, n-octanol/water distribution coefficient, number of hydrogen bond acceptors, number of hydrogen bond donors, total polar surface area, number of rotatable bonds, the number of aromatic rings and the number of alerts for undesirable functional groups) of each compound are presented in Table 2. The molecular weight of the compounds ranges between 78.05 and 518.13 g/mol. The minimum value of the logarithm of the n-octanol/water distribution coefficient (log P) is 0 while the maximum is 11.202. The numbers of hydrogen bond acceptors and donors are maximum of 8 and 3 respectively. Only eight compounds (4-[5-(4-pyridinyl)-1,2,4-oxadiazol-3-yl]-1,2,5-Oxadiazol-3-amine; 2-(2-Hydroxyphenoxy)-1-phenylethanol; 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine; Furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)-; Benzeneethanamine, 4-methoxy-; Decamethyl-Tetrasiloxane; Arsenous acid, tris(trimethylsilyl) ester; Cyclopentadecanone, 2-hydroxy-) with QED values > 0.67 possess desirable drug-likeness and are thus selected for ADMET profiling.
3.2 ADMET profiling
The Absorption, distribution, metabolism, excretion and toxicity (ADME) properties of the eight compounds with desirable QED are summarized in Table 3. The compound, 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine is both a potent inhibitor and substrate of P-glycoprotein while decamethyl-tetrasiloxane and arsenous acid, tris(trimethylsilyl) ester may not be readily absorbed in the human intestine. Furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)- and arsenous acid, tris(trimethylsilyl) ester also show poor Caco-2 permeability. Four compounds (6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine, furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)-; benzeneethanamine, 4-methoxy- and cyclopentadecanone, 2-hydroxy-) may be able to cross the blood–brain barrier. 2-(2-Hydroxyphenoxy)-1-phenylethanol, decamethyl-tetrasiloxane, arsenous acid, tris(trimethylsilyl) ester and cyclopentadecanone, 2-hydroxy-may be tightly bound to plasma protein with 2-(2-Hydroxyphenoxy)-1-phenylethanol and cyclopentadecanone, 2-hydroxy- having free unbound volume of less than 5%. 4-[5-(4-pyridinyl)-1,2,4-oxadiazol-3-yl]-1,2,5-Oxadiazol-3-amine, 2-(2-Hydroxyphenoxy)-1-phenylethanol and decamethyl-tetrasiloxane are likely to exhibit CYP450 inhibition promiscuity as they inhibit more than one isoform of the enzyme. Only furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl), benzeneethanamine, 4-methoxy- and cyclopentadecanone, 2-hydroxy- have a good systemic clearance rate. Although none of the compounds is a potential blocker of hERG (human ether a-go-go) K + channels and may not be mutagenic, 4-[5-(4-pyridinyl)-1,2,4-oxadiazol-3-yl]-1,2,5-Oxadiazol-3-amine and furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl) could cause drug-induced liver injury and lead to hepatocytes toxicity in human. The two compounds, with 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine are potential carcinogens. Only the compounds with no toxic profile were selected for the molecular docking study.
3.3 Homology modelling
LsrK of S. Typhi was modelled using a crystal structure of Escherichia coli K-12 LsrK and HPr complex (PDB id: 5YA0) as a template, having a sequence identity of 83.02% and a GMQE (Global Model Quality Estimate) value of 0.89 with QMEAN (Qualitative Model Energy ANalysis) of 0.88 ± 0.05. Analysis of the Ramachandran plot of the modelled protein for the ϕ and ψ angles of individual amino acid residues revealed that 95.55% of residues were present in the favoured region while 3.64% and 0.81% residues were in the allowed and outlier regions respectively (Fig. 3). Apart from the Ramachandran outliers, the protein also has Rotamer outliers of about 1.01%. The total number of bad angles present in the chain is 78 out of 5,252.
The template for SdiA of S. Typhi was a 3-D crystal structure of E. coli SdiA in complex with 3-oxo-C6-homoserine lactone (PDB id: 4Y15) with a sequence identity of 72.08% and a GMQE value of 0.88 with QMEAN of 0.85 ± 0.05. The Ramachandran plot and the 3-D structure of the model are presented in Fig. 4. Analysis of the Ramachandran plot of the modelled protein for the ϕ and ψ angles of individual amino acid residues revealed that 97.01% and 2.99% residues were present in the favoured and allowed regions respectively. There was no Ramachandran outlier while Rotamer outliers were about 0.48%. The total number of bad angles present in the chain is 16 out of 5398.
3.4 Molecular docking
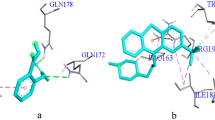
Molecular docking revealed that only two compounds (Benzeneethanamine, 4-methoxy- and Cyclopentadecanone, 2-hydroxy-) have an interaction with each of the four proteins studied (Table 4). The energy of affinities of these two compounds was only lower than that of ciprofloxacin in their interactions with SdiA. For all the proteins, Cyclopentadecanone, 2-hydroxy-has a better (lower) affinity energy than Benzeneethanamine, 4-methoxy-. Analysis of the docking output (Table 5 and Fig. 5) revealed that, except for SdiA, Benzeneethanamine, 4-methoxy-interacts with chain A of each protein with more hydrogen bonds than with any other type of (non-covalent) bonds. Almost all the interactions are with polar amino acids. For Cyclopentadecanone, 2-hydroxy-, analysis of the docking output (Table 6 and Fig. 6) revealed fewer hydrogen bond interactions with each protein. Most of the interactions are with basic or neutral amino acids.
4 Discussion
The process of drug discovery has been made easier with the help of computational drug designing which uses various computational methods to predict interactions between potential drug targets and small molecules to determine the best lead compounds.
Virtual screening of the drug-likeness of small molecules is being estimated using the physicochemical properties of such molecules. Many researchers have come up with different sets of rules regarding the suitability or otherwise of compounds for lead-likeness based on their physicochemical properties, however, such rules are based on a few of those properties. For example, Lipinski's Rule of 5 is based on 4 of those parameters (molecular weight, LogP, Number of hydrogen bond acceptors and donors) [26]. Conversely, QED is a multi-criteria optimization that offers quantitative metrics based on 8 physicochemical parameters and offers a clear-cut distinction between desirable and undesirable compounds (Bickerton et al. [5]). Although many of the compounds studied did not violate more than one of Lipinski's Rule of 5, only eight of them have the 'desirable' value of QED and were thus selected for ADMET profiling. This would ensure that scarce resources are not wasted on molecules which are likely to be abandoned at a later stage of drug design.
In silico prediction ofADMET profiles of small molecules offers the advantage of early detection of poor pharmacokinetic and toxic potentials, avoiding costly late-stage failure in drug development, which is less expensive and time-saving. P-glycoprotein substrate or inhibitor, human intestinal absorption and Caco-2 permeability were used to predict the absorption level of the compounds. Inhibition of P-glycoprotein by 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine, low human intestinal absorption of compounds Decamethyl-Tetrasiloxane and Arsenous acid, tris(trimethylsilyl) ester, as well as low apparent Caco-2 permeability of furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)- and Arsenous acid, tris(trimethylsilyl) ester could lower the bioavailability of these compounds in human. 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine, furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)- and Cyclopentadecanone, 2-hydroxy- could cross the blood–brain barrier into the brain and cause neurological effects. Whether or not the effect will be beneficial or otherwise is a subject of further investigation as some strains of salmonella have been reported to cause post-typhoid neurological complications [24] and drug molecules with the ability to penetrate the BBB would be valuable in such cases [35]. Five isoforms of cytochrome-P450 enzymes (1A2, 3A4, 2C9, 2C19 and 2D6) metabolize about 90% of xenobiotics. Inhibition of these important enzymes can cause clinically important drug-drug interactions that can lead to adverse reactions or treatment failures [27]. 4-[5-(4-pyridinyl)-1,2,4-oxadiazol-3-yl]-1,2,5-Oxadiazol-3-amine, 2-(2-Hydroxyphenoxy)-1-phenylethanol, furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)- and Benzeneethanamine, 4- methoxy- inhibit at least one isoform of cytochrome-P450 and this poses a hindrance to their suitability as drug candidates unless they are improved upon at the optimization stage. Five toxicity parameters were considered for screening of small molecules in this study viz: hERG, H-HT, DILI, Ames and carcinogenicity. Due to the central role played by the liver in human metabolism, human hepatotoxicity and drug-induced liver injury are important parameters to consider in any prospective drug molecule [6]. Carcinogenicity prediction is also an important parameter as it measures the probability that a molecule may induce cancer. Compounds with potential mutagenicity and carcinogenicity are not further considered in drug design [21]. Based on these three parameters. Three of the compounds (4-[5-(4-pyridinyl)-1,2,4-oxadiazol-3-yl]-1,2,5-Oxadiazol-3-amine, 6-Methyl-4-propan-2-on-3-propyl-2, 6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine and Furazan-3-amine,4-(4-methyl-1-piperidylcarbonyl)-) were thus removed from further consideration.
Homology modelling provides a model of 3-D structures of proteins whose experimental 3-D crystal structures are not available in the Protein Data Bank. The reliability of such structures depends on metrics such as the Global Model Quality Estimation (GMQE) and Qualitative Model Energy Analysis (QMEAN) values. The GMQE value ranges between 0 and 1, and the higher the number, the higher the reliability of the predicted structure, while QMEAN < 4.0 shows reliability [29]. With GMQE and QMEAN of 0.89 and 0.88 ± 0.05 respectively for the modelled LsrK, the model is reliable enough for the molecular docking study. The SdiA model is also reliable (GMQE = 0.88, QMEAN = 0.85 ± 0.05). The torsional angles of this protein are good, according to the Ramachandran plot study. As a result, the structures are reliable.
Given the pivotal role played by the QS network in the development of both carrier status and antibiotic resistance in typhoid fever, and indeed other important bacterial infections, QS inhibitors are gaining more popularity as prospective alternatives in combating bacterial diseases (Jiang et al., [23]). Proteins in the QS network of S. Typhi selected for study are LuxS protein (synthesis of AI-2), LsrB (binding and transport of AI-2 into the cell), LsrK (intracellular phosphorylation of AI-2) and SdiA (transcription factor which is the receptor of the processed AI). Although LuxS does not affect the growth of Salmonella spp., it plays a role in the formation of biofilm as it catalyzes a crucial step in the synthesis of AI-2. It also controls the expression of polarization of the bacterial phase variation [33], the catalytic site of LuxS comprises a Zn2+ coordinated by HIS: A54, HIS: A58 and CYS: A126. These amino acids are crucial for its catalysis [20]. Our molecular docking results show that Benzeneethanamine, 4-methoxy- bind HIS: A58 of LuxS with both Pi-Akyl and Pi-Pi T-shaped bonds and this could inhibit the protein. There is also a hydrogen bond at the neighbouring GLUA:57. Also, Benzeneethanamine, 4- methoxy- also interacts with TRY B:95, ALA B:110 and GLY B:118 while Cyclopentadec anone, 2-hydroxy-interacts with ASP A:80, all of which form part of the binding site of SdiA [17]. Better interactions of the compounds with SdiA (− 6.20 kcal/mol for Benzeneethanamine, 4- methoxy- and − 7.70 kcal/mol for Cyclopentadecanone, 2-hydroxy-) relative to ciprofloxacin (− 4.30 kcal/mol) indicate that they fit well into the binding pockets of the protein and makes them promising S. Typhi QS inhibitors as inactivation of the processed AI receptors has been proven to reduce virulence in some pathogenic bacteria [45, 48]. Interestingly Cyclopentadecanone, 2-hydroxy- has been previously reported to inhibit DNA gyrase, another important protein involved in the replication of S. Typhi [2]. Thus, Cyclopentadecanone, 2-hydroxy-, and Benzeneethanamine, 4-methoxy- are being considered for lead optimization.
5 Conclusion
QED, being a multi-criteria optimization, gave a numerical output which made it straightforward in determining the drug-likeness of the phytocompounds studied. Only eight out of seventy-two compounds had drug-likeness desirability (QED > 0.67). Prediction of ADMET profile accords the removal of compounds that could later be withdrawn if made into drugs. This led to discontinuation with three compounds due to predicted human hepatotoxicity, drug-induced liver injury and carcinogenicity. Benzeneethanamine, 4-methoxy- and Cyclopentadecanone, 2-hydroxy- from P. guajava leaves show remarkable interaction with all four QS proteins of S. Typhi studied with negative binding energies. Future research focused on molecular dynamics of the interactions as well as optimization of the two compounds will give more insight into the possibility of exploring them through in vitro and in vivo experiments against the deadly salmonella specie.
Data availability
All data generated during this study are contained in this published article.
References
Adetutu A, Olaniyi TD, Awodugba T, Owoade AO, Olaniyan LWB, Oyekunle SO (2021) In vitro and in vivo activities of Psidium guajava and Azadirachta indica leaf extracts and solvent fractions against Salmonella Typhi. European J Med Plants 32(7):56–71. https://doi.org/10.9734/EJMP/2021/v32i730405
Adetutu A, Olaniyi TD, Owoade AO (2021) GC-MS analysis and in silico assessment of constituents of Psidium guajava leaf extract against DNA gyrase of Salmonella enterica serovar Typhi. Inform Med Unlocked. https://doi.org/10.1016/j.imu.2021.100722
Ashurst, JV, Truong J, Woodbury B (2021) Salmonella Typhi. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK519002/ Accessed 16/06/2022
Borges A, Abreu AC, Dias C, Saavedra MJ, Borges F, Simões M (2016) New Perspectives on the use of phytochemicals as an emergent strategy to control bacterial infections including biofilms. Molecules 21:877. https://doi.org/10.3390/molecules21070877
Bickerton, GR, Paolini GV, Besnard J, Muresan S, Hopkins AL (2012) Quantifying the chemical beauty of drugs. Nat Chem 4(2):90–98. https://doi.org/10.1038/nchem.1243
Bitew M, Desalegn T, Demissie TB, Belayneh A, Endale M, Eswaramoorthy R (2021) Pharmacokinetics and drug-likeness of antidiabetic flavonoids: molecular docking and DFT study. PLoS ONE 16(12):e0260853. https://doi.org/10.1371/journal.pone.0260853
Bueno J (2014) Anti-biofilm drug susceptibility testing methods: Looking for new strategies against resistance mechanism. J Microb Biochem Technol. https://doi.org/10.4172/1948-5948.S3-004
Cactus online translator-(https://cactus.nci.nih.gov/translate/). Accessed 16 July 2021.
Chelvam KK, Chai LC, Thong KL (2014) Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog 6:2. https://doi.org/10.1186/1757-4749-6-2
Choi J, Shin D, Ryu S (2007) Implication of quorum sensing in Salmonella enterica Serovar Typhimurium Virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect Immun 75(10):4885–4890. https://doi.org/10.1128/IAI.01942-06
Choi J, Shin D, Kim M, Park J, Lim S, Park J, Lim S, Ryu S (2012) LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and Flagella Genes. PLoS ONE 7(5):e37059. https://doi.org/10.1371/journal.pone.0037059
dos Santos RM, Costa G, Cerávolo IP, Dias-Souza MV (2020) Antibiofilm potential of Psidium guajava and Passiflora edulis pulp extracts against Staphylococcus aureus, cytotoxicity, and interference on the activity of antimicrobial drugs. Futur J Pharm Sci. https://doi.org/10.1186/s43094-020-00056-8
Dreszer C, Vrouwenvelder J, Paulitsch-Fuchs A, Zwijnenburg A, Kruithof J et al (2013) Hydraulic resistance of biofilms. J Membr Sci 429:436–447
Fauvart M, De Groote VN, Michiels J (2011) Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709
Fu B, Wu Q, Dang M, Bai D, Guo Q, Shen L, Duan K (2017) Inhibition of Pseudomonas aeruginosa biofilm formation by traditional Chinese medicinal herb Herba patriniae. Biomed Res Int 2017(2017):9584703. https://doi.org/10.1155/2017/9584703
Gibani MM, Jones E, Barton A, Jin C, Meek J, Camara S, Galal U, Heinz E, Rosenberg-Hasson Y, Obermoser G, Jones C, Campbell D, Black C, Thomaides-Brears H, Darlow C, Dold C, Silva-Reyes L, Blackwell L, Lara-Tejero M, Jiao X, Stack G, Blohmke CJ, Hill J, Angus B, Dougan G, Galán J, Pollard AJ (2019) Investigation of the role of typhoid toxin in acute typhoid fever in a human challenge model. Nat Med 25(7):1082–1088. https://doi.org/10.1038/s41591-019-0505-4
Gnanendra S, Anusuya S, Natarajan J (2012) Molecular modelling and active site analysis of SdiA homolog, a putative quorum sensor for Salmonella typhimurium pathogenicity reveals specific binding patterns of AHL transcriptional regulators. J Molecular Modeling 18(10):4709–4719. https://doi.org/10.1007/s00894-012-1469-1
González JF, Alberts H, Lee J, Doolittle L, Gunn JS (2018) Biofilm formation protects salmonella from the antibiotic ciprofloxacin In Vitro and In Vivo in the mouse model of chronic carriage. Sci Rep 8:222. https://doi.org/10.1038/s41598-017-18516-2
Gonzalez-Escobedo G, Marshall JM, Gunn JS (2011) Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9(1):9–14. https://doi.org/10.1038/nrmicro2490
Hilgers MT, Ludwig ML (2001) Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. PNAS 98(20):11169–11174. https://doi.org/10.1073/ypnas.191223098
Hsu KH, Su BH, Tu YS, Lin OA, Tseng YJ (2016) Mutagenicity in a molecule: identification of core structural features of mutagenicity using a scaffold analysis. PLoS ONE 11(2):e0148900. https://doi.org/10.1371/journal.pone.0148900
Jesudhasan PR, Soni KA, Cepeda ML, Hume ME, Widmer K, Zhu J, Dowd SE, Pillai SD (2010) Transcriptome analysis of genes controlled by luxS/Autoinducer-2 in Salmonella enterica Serovar Typhimurium. Foodborne Pathog Dis 7(4):339–410. https://doi.org/10.1089/fpd.2009.0372
Jiang Q, Chen J, Yang C, Yin Y, Yao K (2019) Quorum sensing: a prospective therapeutic target for bacterial diseases. Biomed Res Int 2019:2015978. https://doi.org/10.1155/2019/2015978
Kaur A, Chopra K, Kaur IP, Rishi P (2020) Salmonella strain specificity determines post-typhoid central nervous system complications: Intervention by Lactiplantibacillus plantarum at Gut-Brain Axis. Front Microbiol 11:1568. https://doi.org/10.3389/fmicb.2020.01568
Koopman JA, Marshall JM, Bhatiya A, Eguale T, Kwiek JJ, Gunn JS (2015) Inhibition of Salmonella enterica biofilm formation using small-molecule adenosine mimetics. Antimicrob Agents Chemother 59(1):76–84. https://doi.org/10.1128/AAC.03407-14
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26. https://doi.org/10.1016/s0169-409x(00)00129-0
Lynch T, Price A (2007) The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician 76(3):391–396
Millones-Gómez PA, Maurtua-Torres D, Bacilio-Amaranto R, Calla-Poma RD, Requena-Mendizabal MF, Valderrama-Negron AC, Calderon-Miranda MA, Calla-Poma RA, Huauya-Leuyacc ME (2020) Antimicrobial activity and anti-adherent effect of peruvian Psidium guajava (Guava) leaves on a cariogenic biofilm model. J Contemp Dent Pract 21(7):733–740
Oduselu GO, Ajani OO, Ajamma YU, Brors B, Adebiyi E (2019) Homology modelling and molecular docking studies of selected substituted Benzo[d]imidazol-1-yl)methyl)benzimidamide scaffolds on plasmodium falciparum adenylosuccinate lyase receptor. Bioinform Biol Insights 13:1177932219865533. https://doi.org/10.1177/1177932219865533
Olaniyi TD, Awodugba TM, Adetutu A (2021) Ethnobotanical survey and evaluation of anti-salmonella potentials of commonly used plants for typhoid treatment in Ogbomoso, Oyo State. Nigeria J Complement Alterna Med Res 15(1):1–15. https://doi.org/10.9734/JOCAMR/2021/v15i130254
Peele KA, Indira M, Krupanidhi S, Bobby MN, Sanjana N, Babu DJ, Narayana AV, Venkateswarulu TC (2020) In-silico Screening of antibiofilm activity against Acinetobacter baumannii. Acta Sci Biotechnol 1(1):8–10
Pena RT, Blasco L, Ambroa A, González-Pedrajo B, Fernández-García L, López M, Bleriot I, Bou G, García-Contreras R, Wood TK, Tomás M (2019) Relationship between quorum sensing and secretion systems. Front Microbiol 10:1100. https://doi.org/10.3389/fmicb.2019.01100
PubChem https://pubchem.ncbi.nlm.nih.gov/. Accessed 16 July 2021.
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385. https://doi.org/10.1093/nar/gkg520
Shaikh AIA, Prabhakar AT (2021) Typhoid fever and its nervous system involvement. In: Saxena SK, Prakash H (eds) Innate immunity in health and disease. IntechOpen, London
Sharma D, Misba L, Khan AU (2019) Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control 8:76. https://doi.org/10.1186/s13756-019-0533-3
Sholpan A, Lamas A, Cepeda A, Franco CM (2021) Salmonella spp. quorum sensing: an overview from environmental persistence to host cell invasion. AIMS Microbiol. 7(2):238–256. https://doi.org/10.3934/microbiol.2021015
Singh S, Singh SK, Chowdhury I, Singh R (2017) Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol J 11:53–62. https://doi.org/10.2174/1874285801711010053
Soto SM (2013) Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4(223–229):13
Trott O, Olson AJ (2010) AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR (2005) Making 'sense' of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat Rev Microbiol 3:383–396. https://doi.org/10.1038/nrmicro1146
Vijayababu P, Samykannu G, Antonyraj CB, Thomas J, Narayanan S, BasheerAhamed SI, Piramanayagam S (2018) Patulin interference with ATP binding cassette transferring auto inducer −2 in Salmonella typhi and biofilm inhibition via quorum sensing. Inform Med Unlocked 11:9–14. https://doi.org/10.1016/j.imu.2018.02.00
Walawalkar YD, Vaidya Y, Nayak V (2016) Response of Salmonella Typhi to bile-generated oxidative stress: implication of quorum sensing and persister cell populations. Pathog Dis 74(8):ftw090. https://doi.org/10.1093/femspd/ftw090
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Weng L, Yang Y, Zhang Y, Wang L (2014) A new synthetic ligand that activates QscR and blocks antibiotic-tolerant biofilm formation in Pseudomonas aeruginosa. Applied Microbiol Biotechnol 98(6):2565–2572. https://doi.org/10.1007/s00253-013-5420
Wu L, Luo Y (2021) Bacterial quorum-sensing systems and their role in intestinal bacteria-host crosstalk. Front Microbiol 12:611413. https://doi.org/10.3389/fmicb.2021.611413
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49(W1):W5-14. https://doi.org/10.1093/nar/gkab255
Yang Y-X, Xu Z-H, Zhang Y-Q, Tian J, Weng L-X, Wang L-H (2012) A new quorum-sensing inhibitor attenuates virulence and decreases antibiotic resistance in Pseudomonas aeruginosa. J Microbiol 50(6):987–993. https://doi.org/10.1007/s12275-012-2149-7
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by TDO. The first draft of the manuscript was written by TDO and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Olaniyi, T.D., Adetutu, A. In silico anti-quorum sensing activities of phytocompounds of Psidium guajava in Salmonella enterica serovar Typhi. J.Umm Al-Qura Univ. Appll. Sci. 9, 142–156 (2023). https://doi.org/10.1007/s43994-023-00029-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43994-023-00029-6