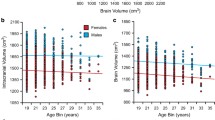
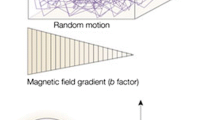
Abstract
Imaging-derived phenotypes (IDPs) have been increasingly used in population-based cohort studies in recent years. As widely reported, magnetic resonance imaging (MRI) is an important imaging modality for assessing the anatomical structure and function of the brain with high resolution and excellent soft-tissue contrast. The purpose of this article was to describe the imaging protocol of the brain MRI in the China Phenobank Project (CHPP). Each participant underwent a 30-min brain MRI scan as part of a 2-h whole-body imaging protocol in CHPP. The brain imaging sequences included T1-magnetization that prepared rapid gradient echo, T2 fluid-attenuated inversion-recovery, magnetic resonance angiography, diffusion MRI, and resting-state functional MRI. The detailed descriptions of image acquisition, interpretation, and post-processing were provided in this article. The measured IDPs included volumes of brain subregions, cerebral vessel geometrical parameters, microstructural tracts, and function connectivity metrics.








Similar content being viewed by others
Data availability
The data supported the protocols of this study are available on request from the corresponding author.
Abbreviations
- AAL:
-
Automated anatomical labeling
- AD:
-
Axial diffusivity
- ADC:
-
Apparent diffusion coefficient
- ALFF:
-
Amplitude of low-frequency fluctuation
- CAT:
-
Computational anatomy toolbox
- CHPP:
-
China Phenobank Project
- CSF:
-
Cerebrospinal fluid
- DC:
-
Degree centrality
- DWI:
-
Diffusion weighted imaging
- DTI:
-
Diffusion tensor imaging
- EPI:
-
Echo-echo planar imaging
- FA:
-
Fractional anisotropy
- FC:
-
Functional connectivity
- FLAIR:
-
Fluid-attenuated inversion-recovery
- GRE:
-
Gradient recalled echo
- IDP:
-
Imaging-derived phenotype
- MD:
-
Mean diffusivity
- MPRAGE:
-
Magnetization prepared rapid gradient echo
- MRI:
-
Magnetic resonance imaging
- RD:
-
Radial diffusivity
- ReHo:
-
Regional homogeneity
- rfMRI:
-
Resting-state functional magnetic resonance imaging
- ROI:
-
Region of interest
- SAG:
-
Sagittal
- SE-EPI:
-
Spin echo-echo planar imaging
- T 1w:
-
T1-Weighted
- T 2-FLAIR:
-
T2-Weighted fluid attenuated inversion recovery
- TBSS:
-
Tract‑based spatial statistics
- TOF-MRA:
-
Time of flight-magnetic resonance angiography
- TRA:
-
Transverse
- TSE:
-
Turbo spin echo
References
Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E, Vidaurre D, Webster M, McCarthy P, Rorden C, Daducci A, Alexander DC, Zhang H, Dragonu I, Matthews PM, Miller KL, Smith SM (2018) Image processing and Quality control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166:400–424. https://doi.org/10.1016/j.neuroimage.2017.10.034
Anand SS, Tu JV, Awadalla P, Black S, Boileau C, Busseuil D, Desai D, Després JP, de Souza RJ, Dummer T, Jacquemont S, Knoppers B, Larose E, Lear SA, Marcotte F, Moody AR, Parker L, Poirier P, Robson PJ, Smith EE, Spinelli JJ, Tardif JC, Teo KK, Tusevljak N, Friedrich MG (2016) Rationale, design, and methods for Canadian alliance for healthy hearts and minds cohort study (CAHHM)—a Pan Canadian cohort study. BMC Public Health 16:650. https://doi.org/10.1186/s12889-016-3310-8
Andersson C, Johnson AD, Benjamin EJ, Levy D, Vasan RS (2019) 70-year legacy of the Framingham Heart Study. Nat Rev Cardiol 16(11):687–698. https://doi.org/10.1038/s41569-019-0202-5
Andersson J, Smith S, Jenkinson M (2008) FNIRT-FMRIB’s non-linear image registration tool. In: 14th annual meeting of the organization for human brain mapping (OHBM), pp 15–19
Ashburner J (2012) SPM: a history. Neuroimage 62(2):791–800. https://doi.org/10.1016/j.neuroimage.2011.10.025
Bamberg F, Kauczor HU, Weckbach S, Schlett CL, Forsting M, Ladd SC, Greiser KH, Weber MA, Schulz-Menger J, Niendorf T, Pischon T, Caspers S, Amunts K, Berger K, Bülow R, Hosten N, Hegenscheid K, Kröncke T, Linseisen J, Günther M, Hirsch JG, Köhn A, Hendel T, Wichmann HE, Schmidt B, Jöckel KH, Hoffmann W, Kaaks R, Reiser MF, Völzke H (2015) Whole-body MR imaging in the German national cohort: rationale, design, and technical background. Radiology 277(1):206–220. https://doi.org/10.1148/radiol.2015142272
Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995) Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34(4):537–541. https://doi.org/10.1002/mrm.1910340409
Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA (2009) Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci 29(6):1860–1873. https://doi.org/10.1523/jneurosci.5062-08.2009
Chau W, McIntosh AR (2005) The Talairach coordinate of a point in the MNI space: how to interpret it. Neuroimage 25(2):408–416. https://doi.org/10.1016/j.neuroimage.2004.12.007
Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002) Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33(3):341–355. https://doi.org/10.1016/s0896-6273(02)00569-x
Frangi AF, Niesen WJ, Vincken KL, et al (1998) Multiscale vessel enhancement filtering. In: In medical image computing and computer-assisted intervention—MICCAI'98. Springer, Berlin, pp 130–137. https://doi.org/10.1007/BFb0056195
Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R (1996) Movement-related effects in fMRI time-series. Magn Reson Med 35(3):346–355. https://doi.org/10.1002/mrm.1910350312
Garrison MM, Ward TM (2019) 0746 two year follow-up of the SHIP (Sleep Health In Preschoolers) Randomized Trial: trajectories of change. Sleep 42(Suppl_1):A299–A300. https://doi.org/10.1093/sleep/zsz067.744
Glasser MF, Smith SM, Marcus DS, Andersson JL, Auerbach EJ, Behrens TE, Coalson TS, Harms MP, Jenkinson M, Moeller S, Robinson EC, Sotiropoulos SN, Xu J, Yacoub E, Ugurbil K, Van Essen DC (2016) The Human Connectome Project’s neuroimaging approach. Nat Neurosci 19(9):1175–1187. https://doi.org/10.1038/nn.4361
Gong W, Beckmann CF, Smith SM (2021) Phenotype discovery from population brain imaging. Med Image Anal 71:102050. https://doi.org/10.1016/j.media.2021.102050
Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YOJSd (2016) The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3(1):1–9. https://doi.org/10.1038/sdata.2016.44
Griffanti L, Zamboni G, Khan A, Li L, Bonifacio G, Sundaresan V, Schulz UG, Kuker W, Battaglini M, Rothwell PM, Jenkinson M (2016) BIANCA (Brain Intensity AbNormality Classification Algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage 141:191–205. https://doi.org/10.1016/j.neuroimage.2016.07.018
He Q, Zhang C, Liu DC (2015) Nonlinear image enhancement by self-adaptive sigmoid function. Int J Signal Process Image Process Pattern Recogn 8(11):319–328. https://doi.org/10.14257/ijsip.2015.8.11.29
Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K (2015) A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage 115:117–137. https://doi.org/10.1016/j.neuroimage.2015.04.042
Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM (2012) FSL. Neuroimage 62(2):782–790. https://doi.org/10.1016/j.neuroimage.2011.09.015
Jiang J, Liu T, Zhu W, Koncz R, Liu H, Lee T, Sachdev PS (2018) UBO Detector-A cluster-based, fully automated pipeline for extracting white matter hyperintensities. Neuroimage 174:539–549. https://doi.org/10.1016/j.neuroimage.2018.03.050
Littlejohns TJ, Holliday J, Gibson LM, Garratt S, Oesingmann N, Alfaro-Almagro F, Bell JD, Boultwood C, Collins R, Conroy MC, Crabtree N, Doherty N, Frangi AF, Harvey NC, Leeson P, Miller KL, Neubauer S, Petersen SE, Sellors J, Sheard S, Smith SM, Sudlow CLM, Matthews PM, Allen NE (2020) The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat Commun 11(1):2624. https://doi.org/10.1038/s41467-020-15948-9
Petersen SE, Matthews PM, Bamberg F, Bluemke DA, Francis JM, Friedrich MG, Leeson P, Nagel E, Plein S, Rademakers FE, Young AA, Garratt S, Peakman T, Sellors J, Collins R, Neubauer S (2013) Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank—rationale, challenges and approaches. J Cardiovasc Magn Reson 15(1):46. https://doi.org/10.1186/1532-429x-15-46
Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, Scholz M, Sargurupremraj M, Jahanshad N, Roshchupkin GV, Smith AV, Bis JC, Jian X, Luciano M, Hofer E, Teumer A, van der Lee SJ, Yang J, Yanek LR, Lee TV, Li S, Hu Y, Koh JY, Eicher JD, Desrivières S, Arias-Vasquez A, Chauhan G, Athanasiu L, Rentería ME, Kim S, Hoehn D, Armstrong NJ, Chen Q, Holmes AJ, den Braber A, Kloszewska I, Andersson M, Espeseth T, Grimm O, Abramovic L, Alhusaini S, Milaneschi Y, Papmeyer M, Axelsson T, Ehrlich S, Roiz-Santiañez R, Kraemer B, Håberg AK, Jones HJ, Pike GB, Stein DJ, Stevens A, Bralten J, Vernooij MW, Harris TB, Filippi I, Witte AV, Guadalupe T, Wittfeld K, Mosley TH, Becker JT, Doan NT, Hagenaars SP, Saba Y, Cuellar-Partida G, Amin N, Hilal S, Nho K, Mirza-Schreiber N, Arfanakis K, Becker DM, Ames D, Goldman AL, Lee PH, Boomsma DI, Lovestone S, Giddaluru S, Le Hellard S, Mattheisen M, Bohlken MM, Kasperaviciute D, Schmaal L, Lawrie SM, Agartz I, Walton E, Tordesillas-Gutierrez D, Davies GE, Shin J, Ipser JC, Vinke LN, Hoogman M, Jia T, Burkhardt R, Klein M, Crivello F, Janowitz D, Carmichael O, Haukvik UK, Aribisala BS, Schmidt H, Strike LT, Cheng CY, Risacher SL, Pütz B, Fleischman DA, Assareh AA, Mattay VS, Buckner RL, Mecocci P, Dale AM, Cichon S, Boks MP, Matarin M, Penninx B, Calhoun VD, Chakravarty MM, Marquand AF, Macare C, Kharabian Masouleh S, Oosterlaan J, Amouyel P, Hegenscheid K, Rotter JI, Schork AJ, Liewald DCM, de Zubicaray GI, Wong TY, Shen L, Sämann PG, Brodaty H, Roffman JL, de Geus EJC, Tsolaki M, Erk S, van Eijk KR, Cavalleri GL, van der Wee NJA, McIntosh AM, Gollub RL, Bulayeva KB, Bernard M, Richards JS, Himali JJ, Loeffler M, Rommelse N, Hoffmann W, Westlye LT, Valdés Hernández MC, Hansell NK, van Erp TGM, Wolf C, Kwok JBJ, Vellas B, Heinz A, Olde Loohuis LM, Delanty N, Ho BC, Ching CRK, Shumskaya E, Singh B, Hofman A, van der Meer D, Homuth G, Psaty BM, Bastin ME, Montgomery GW, Foroud TM, Reppermund S, Hottenga JJ, Simmons A, Meyer-Lindenberg A, Cahn W, Whelan CD, van Donkelaar MMJ, Yang Q, Hosten N, Green RC, Thalamuthu A, Mohnke S, Hulshoff Pol HE, Lin H, Jack CR, Jr., Schofield PR, Mühleisen TW, Maillard P, Potkin SG, Wen W, Fletcher E, Toga AW, Gruber O, Huentelman M, Davey Smith G, Launer LJ, Nyberg L, Jönsson EG, Crespo-Facorro B, Koen N, Greve DN, Uitterlinden AG, Weinberger DR, Steen VM, Fedko IO, Groenewold NA, Niessen WJ, Toro R, Tzourio C, Longstreth WT, Jr., Ikram MK, Smoller JW, van Tol MJ, Sussmann JE, Paus T, Lemaître H, Schroeter ML, Mazoyer B, Andreassen OA, Holsboer F, Depondt C, Veltman DJ, Turner JA, Pausova Z, Schumann G, van Rooij D, Djurovic S, Deary IJ, McMahon KL, Müller-Myhsok B, Brouwer RM, Soininen H, Pandolfo M, Wassink TH, Cheung JW, Wolfers T, Martinot JL, Zwiers MP, Nauck M, Melle I, Martin NG, Kanai R, Westman E, Kahn RS, Sisodiya SM, White T, Saremi A, van Bokhoven H, Brunner HG, Völzke H, Wright MJ, van 't Ent D, Nöthen MM, Ophoff RA, Buitelaar JK, Fernández G, Sachdev PS, Rietschel M, van Haren NEM, Fisher SE, Beiser AS, Francks C, Saykin AJ, Mather KA, Romanczuk-Seiferth N, Hartman CA, DeStefano AL, Heslenfeld DJ, Weiner MW, Walter H, Hoekstra PJ, Nyquist PA, Franke B, Bennett DA, Grabe HJ, Johnson AD, Chen C, van Duijn CM, Lopez OL, Fornage M, Wardlaw JM, Schmidt R, DeCarli C, De Jager PL, Villringer A, Debette S, Gudnason V, Medland SE, Shulman JM, Thompson PM, Seshadri S, Ikram MA (2019) Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet 51(11):1624–1636. https://doi.org/10.1038/s41588-019-0511-y
Sem F (2012) Automatic segmentation of nucleus accumbens. Dissertation, Universitat Politècnica de Catalunya. http://hdl.handle.net/2099.1/16445
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155. https://doi.org/10.1002/hbm.10062
Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE (2006) Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31(4):1487–1505. https://doi.org/10.1016/j.neuroimage.2006.02.024
Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, Elliott LT (2021) An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci 24(5):737–745. https://doi.org/10.1038/s41593-021-00826-4
Suever J (2011) DICOM Sort web. A free and flexible sorting utility. https://dicomsort.com/. Accessed 14 May, 2022
Tian Y, Chen HB, Ma XX, Li SH, Li CM, Wu SH, Liu FZ, Du Y, Li K, Su W (2022) Aberrant volume-wise and voxel-wise concordance among dynamic intrinsic brain activity indices in Parkinson’s disease: a resting-state fMRI study. Front Aging Neurosci 14:814893. https://doi.org/10.3389/fnagi.2022.814893
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Étard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289. https://doi.org/10.1006/nimg.2001.0978
Victor RG, Haley RW, Willett DL, Peshock RM, Vaeth PC, Leonard D, Basit M, Cooper RS, Iannacchione VG, Visscher WA, Staab JM, Hobbs HH (2004) The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol 93(12):1473–1480. https://doi.org/10.1016/j.amjcard.2004.02.058
Zang Y, Jiang T, Lu Y, He Y, Tian L (2004) Regional homogeneity approach to fMRI data analysis. Neuroimage 22(1):394–400. https://doi.org/10.1016/j.neuroimage.2003.12.030
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20(1):45–57. https://doi.org/10.1109/42.906424
Zhang J, Liu DQ, Qian S, Qu X, Zhang P, Ding N, Zang YF (2022) The neural correlates of amplitude of low-frequency fluctuation: a multimodal resting-state MEG and fMRI-EEG study. Cereb Cortex. https://doi.org/10.1093/cercor/bhac124
Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP (2012) Network centrality in the human functional connectome. Cereb Cortex 22(8):1862–1875. https://doi.org/10.1093/cercor/bhr269
Acknowledgements
The data and samples used for this protocol were obtained from CHPP. We would like to thank the CHPP participants and coordinators for their contribution to this dataset.
Funding
This study was funded by the Shanghai Municipal Science and Technology Major Project (No. 2017SHZDZX01).
Author information
Authors and Affiliations
Contributions
CW, MT, HW: concept and design, data interpretation and analysis, supervision, drafting, revision and approval of final manuscript. ZS, YL, NH, YG, WC, JZ, JL: data interpretation and analysis, drafting, revision and approval of final manuscript. XX, XK, SQ, LX, LL, YW, NZ, JT, XH, WC: data collection, data analysis, drafting, revision and approval of final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was in agreement with the ethical guidelines of the 1975 Declaration of Helsinki and approved by the institutional review board of Fudan University. Informed consent was obtained from each subject.
Consent for publication
Not applicable.
Competing interests
MT is the Editorial Board Member of Phenomics, and she was not involved in reviewing this paper.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, C., Shi, Z., Li, Y. et al. Protocol for Brain Magnetic Resonance Imaging and Extraction of Imaging-Derived Phenotypes from the China Phenobank Project. Phenomics 3, 642–656 (2023). https://doi.org/10.1007/s43657-022-00083-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-022-00083-w