Abstract
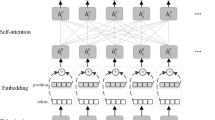
The named entity recognition (NER) is a method for locating references to rigid designators in text that fall into well-established semantic categories like person, place, organisation, etc., Many natural language e applications, like summarization of text, question–answer models and machine translation, always include NER at their core. Early NER systems were quite successful in reaching high performance at the expense of using human engineers to create features and rules that were particular to a certain domain. Currently, the biomedical data is soaring expeditiously and extracting the useful information can help to facilitate the appropriate diagnosis. Therefore, these systems are widely adopted in biomedical domain. However, the traditional rule-based, dictionary based and machine learning based methods suffer from computational complexity and out-of-vocabulary (OOV)issues Deep learning has recently been used in NER systems, achieving the state-of-the-art outcome. The present work proposes a novel deep learning based approach which uses Bidirectional Long Short Term (BiLSTM), Bidirectional Encoder Representation (BERT) and Conditional Random Field mode (CRF) model along with transfer learning and multi-tasking model to solve the OOV problem in biomedical domain. The transfer learning architecture uses shared and task specific layers to achieve the multi-task transfer learning task. The shared layer consists of lexicon encoder and transformer encoder followed by embedding vectors. Finally, we define a training loss function based on the BERT model. The proposed Multi-task TLBBC approach is compared with numerous prevailing methods. The proposed Multi-task TLBBC approach realizes average accuracy as 97.30%, 97.20%, 96.80% and 97.50% for NCBI, BC5CDR, JNLPBA, and s800 dataset, respectively.




Similar content being viewed by others
Data Availability
The dataset generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. J Big Data. 2019;6(1):1–25.
Lou Z, Wang L, Jiang K, Wei Z, Shen G. Reviews of wearable healthcare systems: materials, devices and system integration. Mater Sci Eng R Rep. 2020;140: 100523.
Pooja H, Jagadeesh MP. A collective study of data mining techniques for the big health data available from the electronic health records. In: 2019 1st International conference on advanced technologies in intelligent control, environment, computing & communication engineering (ICATIECE), Bangalore, India; 2019. p. 51–55. https://doi.org/10.1109/ICATIECE45860.2019.9063623.
Zilbermint M. Diabetes and climate change. J Commun Hosp Intern Med Perspect. 2020;10(5):409–12.
Sung M, Jeong M, Choi Y, Kim D, Lee J, Kang J. BERN2: an advanced neural biomedical named entity recognition and normalization tool. Bioinformatics. 2022;38(20):4837–9.
Wen Y, Fan C, Chen G, Chen X, Chen M. A survey on named entity recognition. In: Communications, signal processing, and systems: proceedings of the 8th international conference on communications, signal processing, and systems. Springer Singapore; 2020. p. 1803–1810.
Giorgi JM, Bader GD. Towards reliable named entity recognition in the biomedical domain. Bioinformatics. 2020;36(1):280–6.
Song B, Li F, Liu Y, Zeng X. Deep learning methods for biomedical named entity recognition a survey and qualitative comparison. Brief Bioinf. 2021;22(6):bbab282.
Naseem U, Musial K, Eklund P, Prasad M. Biomedical named-entity recognition by hierarchically fusing biobert representations and deep contextual-level word-embedding. In: 2020 International joint conference on neural networks (IJCNN). IEEE; 2020, July. p. 1–8.
Eftimov T, Koroušić Seljak B, Korošec P. A rule-based named-entity recognition method for knowledge extraction of evidence-based dietary recommendations. PLoS One. 2017;12(6): e0179488.
Asghari M, Sierra-Sosa D, Elmaghraby AS. BINER: A low-cost biomedical named entity recognition. Inf Sci. 2022;602:184–200.
Cho M, Ha J, Park C, Park S. Combinatorial feature embedding based on CNN and LSTM for biomedical named entity recognition. J Biomed Inform. 2020;103: 103381.
Naseem U, Khushi M, Reddy V, Rajendran S, Razzak I, Kim J. Bioalbert: a simple and effective pre-trained language model for biomedical named entity recognition. In: 2021 International joint conference on neural networks (IJCNN). IEEE; 2021, July. p. 1–7.
Hong SK, Lee JG. DTranNER: biomedical named entity recognition with deep learning-based label-label transition model. BMC Bioinform. 2020;21:1–11.
Ning G, Bai Y. Biomedical named entity recognition based on Glove-BLSTM-CRF model. J Comput Methods Sci Eng. 2021;21(1):125–33.
Wei H, Gao M, Zhou A, Chen F, Qu W, Wang C, Lu M. Named entity recognition from biomedical texts using a fusion attention-based BiLSTM-CRF. IEEE Access. 2019;7:73627–36.
Çelikmasat G, Aktürk ME, Ertunç YE, Issifu AM, Ganiz MC. Biomedical named entity recognition using transformers with biLSTM+ CRF and graph convolutional neural networks. In: 2022 International conference on innovations in intelligent systems and applications (INISTA). IEEE; 2022, August. p. 1–6.
Zhang Z, Chen AL. Biomedical named entity recognition with the combined feature attention and fully-shared multi-task learning. BMC Bioinform. 2022;23(1):1–21.
Khan MR, Ziyadi M, Abdel Hady M (2020) Mt-bioner: multi-task learning for biomedical named entity recognition using deep bidirectional transformers. arXiv preprint arXiv:2001.08904.
Harnoune A, Rhanoui M, Mikram M, Yousfi S, Elkaimbillah Z, El Asri B. BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput Methods Programs Biomed Update. 2021;1: 100042.
Acknowledgements
The authors acknowledged the JSS Academy of Technical Education, Bengaluru, affiliated to VTU Belagavi, India for supporting the research work by providing the facilities.
Funding
No funding received for this research.
Author information
Authors and Affiliations
Contributions
The dedicated efforts and valuable contributions of all authors involved enabled this collaborative work, significantly enriching the study's outcome through their collective input.
Corresponding author
Ethics declarations
Conflict of Interest
No conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Advances in Computational Approaches for Image Processing, Wireless Networks, Cloud Applications and Network Security” guest edited by P. Raviraj, Maode Ma and Roopashree H R.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pooja, H., Jagadeesh, M.P.P. A Deep Learning Based Approach for Biomedical Named Entity Recognition Using Multitasking Transfer Learning with BiLSTM, BERT and CRF. SN COMPUT. SCI. 5, 482 (2024). https://doi.org/10.1007/s42979-024-02835-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02835-z