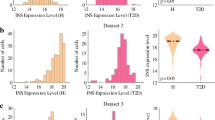
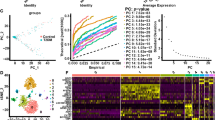
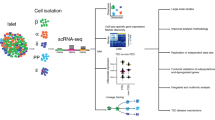
Abstract
Diabetes is a critical disease and is crucial to personage agility. Type 2 Diabetes (T2D) accounts for 92% of epithetical cases. This paper proposes an optimized type 2 diabetes detection model using mixed single-cell RNA sequencing (scRNA-seq) technology. Diabetes is a chronic metabolic disorder affecting millions of people worldwide. Early detection of the disease can greatly improve treatment outcomes, but current diagnostic methods have limitations. Our proposed model integrates scRNA-seq data from both human pancreatic beta cells to identify gene expression patterns associated with diabetes. Our study shows that the proposed model is highly accurate in identifying diabetes, achieving an area under the curve (AUC) of 0.98. We employed an optimized model to improve the detection of diabetes at an early stage, leading to better treatment outcomes and an improved quality of life for patients. We initially incorporated optimal features from the dataset using the Monte Carlo (MC) feature selection method. This method helped us to estimate the relative importance (RI) score of each gene or feature, which is then used to rank the features. Further, we proposed an optimized deep belief network (ODBN) as a classification model to classify T2D and non-diabetes. To improve the performance of ODBN, an adaptive chimp optimization algorithm (AChOA) is introduced to optimize the weight parameters and achieved a performance accuracy of 96.57%.







Similar content being viewed by others
References
Georga EI, Protopappas VC, Ardigo D, Marina M, Zavaroni I, Polyzos D, Fotiadis DI. Multivariate prediction of subcutaneous glucose concentration in type 1 diabetes patients based on support vector regression. IEEE J Biomed Health Inform. 2012;17(1):71–81.
Owens C, Zisser H, Jovanovic L, Srinivasan B, Bonvin D. Run-to-run control of blood glucose concentrations for people with type 1 diabetes mellitus. IEEE Trans Biomed Eng. 2006;53(6):996–1005.
Lee BJ, Kim JY. Identification of type 2 diabetes risk factors using phenotypes consisting of anthropometry and triglycerides based on machine learning. IEEE J Biomed Health Inform. 2015;20(1):39–46.
Cracchiolo M, Sacramento JF, Mazzoni A, Panarese A, Carpaneto J, Conde SV, Micera S. Decoding neural metabolic markers from the carotid sinus nerve in a type 2 diabetes model. IEEE Trans Neural Syst Rehabil Eng. 2019;27(10):2034–43.
Ali O. Genetics of type 2 diabetes. World J Diabetes. 2013;4(4):114.
Gao HX, Regier EE, Close KL. International diabetes federation world diabetes congress. J Diabetes. 2015;8(3):300–2.
Pandey A, Chawla S, Guchhait P. Type-2 diabetes: current understanding and future perspectives. IUBMB Life. 2015;67(7):506–13.
Yabe D, Seino Y, Fukushima M, Seino S. β cell dysfunction versus insulin resistance in the pathogenesis of type 2 diabetes in East Asians. Curr Diabetes Rep. 2015;15:1–9.
Adua E, Kolog EA, Afrifa-Yamoah E, Amankwah B, Obirikorang C, Anto EO, Acheampong E, Wang W, Tetteh AY. Predictive model and feature importance for early detection of type II diabetes mellitus. Transl Med Commun. 2021;6(1):1–5.
Syed AH, Khan T. Machine learning-based application for predicting risk of type 2 diabetes mellitus (T2DM) in Saudi Arabia: a retrospective cross-sectional study. IEEE Access. 2020;8:199539–61.
Nuankaew P, Chaising S, Temdee P. Average weighted objective distance-based method for type 2 diabetes prediction. IEEE Access. 2021;9:137015–28.
Kazerouni F, Bayani A, Asadi F, Saeidi L, Parvizi N, Mansoori Z. Type2 diabetes mellitus prediction using data mining algorithms based on the long-noncoding RNAs expression: a comparison of four data mining approaches. BMC Bioinform. 2020;21:1–3.
Wong WK, Thorat V, Joglekar MV, Dong CX, Lee H, Chew YV, Bhave A, Hawthorne WJ, Engin F, Pant A, Dalgaard LT. Analysis of half a billion datapoints across ten machine-learning algorithms identifies key elements associated with insulin transcription in human pancreatic islet cells. Front Endocrinol. 2022;13: 853863.
Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, Gromada J. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24(4):608–15.
Dramiński M, Rada-Iglesias A, Enroth S, Wadelius C, Koronacki J, Komorowski J. Monte Carlo feature selection for supervised classification. Bioinformatics. 2008;24(1):110–7.
Ngara M, Wierup N. Lessons from single-cell RNA sequencing of human islets. Diabetologia. 2022;65(8):1241–50.
Li D. Statistical methods for rna sequencing data analysis. Exon Publ. 2019;31:85–99.
Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11(7):740–2.
Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16(1):1–3.
Korthauer KD, Chu LF, Newton MA, Li Y, Thomson J, Stewart R, Kendziorski C. A statistical approach for identifying differential distributions in single-cell RNA-seq experiments. Genome Biol. 2016;17:1–5.
Miao Z, Deng K, Wang X, Zhang X. DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics. 2018;34(18):3223–4.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6.
Guo M, Wang H, Potter SS, Whitsett JA, Xu Y. SINCERA: a pipeline for single-cell RNA-Seq profiling analysis. PLoS Comput Biol. 2015;11(11): e1004575.
Delmans M, Hemberg M. Discrete distributional differential expression (D 3 E)-a tool for gene expression analysis of single-cell RNA-seq data. BMC Bioinform. 2016;17:1–3.
Ntranos V, Yi L, Melsted P, Pachter L. A discriminative learning approach to differential expression analysis for single-cell RNA-seq. Nat Methods. 2019;16(2):163–6.
Prabhu P, Selvabharathi S (2019) Deep belief neural network model for prediction of diabetes mellitus. In: 2019 3rd international conference on imaging, signal processing and communication (ICISPC). IEEE, pp. 138–142
Khishe M, Mosavi MR. Chimp optimization algorithm. Expert Syst Appl. 2020;149: 113338.
Balzer MS, Ma Z, Zhou J, Abedini A, Susztak K. How to get started with single cell RNA sequencing data analysis. J Am Soc Nephrol. 2021;32(6):1279–92.
Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y. Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med. 2022;12(3): e694.
Li Z, Pan X, Cai YD. Identification of type 2 diabetes biomarkers from mixed single-cell sequencing data with feature selection methods. Front Bioeng Biotechnol. 2022;10: 890901.
Avrahami D, Wang YJ, Schug J, Feleke E, Gao L, Liu C, Naji A, Glaser B, Kaestner KH. Single-cell transcriptomics of human islet ontogeny defines the molecular basis of β-cell dedifferentiation in T2D. Mol Metab. 2020;42: 101057.
Qadir MM, Álvarez-Cubela S, Klein D, van Dijk J, Muñiz-Anquela R, Moreno-Hernández YB, Lanzoni G, Sadiq S, Navarro-Rubio B, García MT, Díaz Á. Single-cell resolution analysis of the human pancreatic ductal progenitor cell niche. Proc Natl Acad Sci. 2020;117(20):10876–87.
Van Gurp L, Fodoulian L, Oropeza D, Furuyama K, Bru-Tari E, Vu AN, Kaddis JS, Rodríguez I, Thorel F, Herrera PL. Generation of human islet cell type-specific identity genesets. Nat Commun. 2022;13(1):2020.
Wang T, Nabavi S. SigEMD: A powerful method for differential gene expression analysis in single-cell RNA sequencing data. Methods. 2018;145:25–32.
Lindqvist A, Shcherbina L, Prasad RB, Miskelly MG, Abels M, Martínez-Lopéz JA, Fred RG, Nergård BJ, Hedenbro J, Groop L, Hjerling-Leffler J. Ghrelin suppresses insulin secretion in human islets and type 2 diabetes patients have diminished islet ghrelin cell number and lower plasma ghrelin levels. Mol Cell Endocrinol. 2020;511: 110835.
Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–14.
Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks GJ, Larsson AJ, Faridani OR, Sandberg R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38(6):708–14.
Groen N, Leenders F, Mahfouz A, Munoz-Garcia A, Muraro MJ, de Graaf N, Rabelink T, Hoeben R, van Oudenaarden A, Zaldumbide A, Reinders MJ. Single-cell transcriptomics links loss of human pancreatic β-cell identity to ER stress. Cells. 2021;10(12):3585.
Nadesh RK, Arivuselvan K. Type 2: diabetes mellitus prediction using deep neural networks classifier. Int J Cogn Comput Eng. 2020;1:55–61.
Basile G, Kahraman S, Dirice E, Pan H, Dreyfuss JM, Kulkarni RN. Using single-nucleus RNA-sequencing to interrogate transcriptomic profiles of archived human pancreatic islets. Genome Med. 2021;13(1):1–7.
Tonyan ZN, Nasykhova YA, Mikhailova AA, Glotov AS. MicroRNAs as potential biomarkers of type 2 diabetes mellitus. Russ J Genet. 2021;57:764–77.
Squair JW, Gautier M, Kathe C, Anderson MA, James ND, Hutson TH, Hudelle R, Qaiser T, Matson KJ, Barraud Q, Levine AJ. Confronting false discoveries in single-cell differential expression. Nat Commun. 2021;12(1):5692.
Lin Y, Li Y, Huang X, Liu L, Wei H, Zou X. Analysis of diabetes clinical data based on recurrent neural networks. Comput Intell Neurosci. 2022. https://doi.org/10.1155/2022/4755728.
Madan P, Singh V, Chaudhari V, Albagory Y, Dumka A, Singh R, Gehlot A, Rashid M, Alshamrani SS, AlGhamdi AS. An optimization-based diabetes prediction model using CNN and Bi-directional LSTM in real-time environment. Appl Sci. 2022;12(8):3989.
Chen K, Zhang J, Huang Y, Tian X, Yang Y, Dong A. Single-cell RNA-seq transcriptomic landscape of human and mouse islets and pathological alterations of diabetes. Nat Commun. 2022;25: 105366.
Tonyan ZN, Nasykhova YA, Danilova MM, Barbitoff YA, Changalidi AI, Mikhailova AA, Glotov AS. Overview of transcriptomic research on type 2 diabetes: challenges and perspectives. Nat Commun. 2022;7:1176.
Taylor HJ, Hung Y-H, Narisu N, Collins FS, Taylor DL. Human pancreatic islet microRNAs implicated in diabetes and related traits by large-scale genetic analysis. Proc Natl Acad Sci U S A. 2023;120(7): e2206797120.
Rubio-Navarro A, Gómez-Banoy N, Stoll L, et al. A beta cell subset with enhanced insulin secretion and glucose metabolism is reduced in type 2 diabetes. Nat Cell Biol. 2023;25(4):565–78.
Olaniru OE, Kadolsky U, Kannambath S, Vaikkinen H, Fung K, Dhami P, Persaud SJ. Single-cell transcriptomic and spatial landscapes of the developing human pancreas. Cell Metab. 2023;35:184–99.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare we have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Advances in Computational Intelligence for Artificial Intelligence, Machine Learning, Internet of Things and Data Analytics” guest edited by S. Meenakshi Sundaram, Young Lee, and Gururaj K S.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Padmaja, K., Mukhopadhyay, D. A Model for Detecting Type 2 Diabetes Using Mixed Single-Cell RNA Sequencing with Optimized Data. SN COMPUT. SCI. 4, 768 (2023). https://doi.org/10.1007/s42979-023-02215-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-023-02215-z