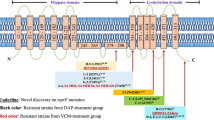
Abstract
Strain C1 was successfully isolated from an immunosuppressed patient with persistent bacteremia, who had not previously been exposed to glycopeptide antibiotics. This strain was found to be a heterogeneous vancomycin intermediate–resistant Staphylococcus aureus (hVISA). It is noteworthy that, following a brief period of vancomycin treatment, strains C6, C8, and C9, which were obtained from blood and other body parts, exhibited a significant reduction in heterogeneity as determined by population analysis profile–area under the curve (PAP-AUC) detection. Genotyping analysis revealed that these bacterial strains belonged to the same SCCmecIVa-ST59-t437-agrI genotype and shared the same virulome and resistome. In this study, a comparative genomics analysis was conducted between strain C1 and strain N315 to identify potential hVISA-associated mutations. Ultimately, a total of 205 mutation sites in 19 candidate genes, likely associated with the hVISA phenotype, were identified.




Similar content being viewed by others
Data Availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S et al (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673. https://doi.org/10.1016/S0140-6736(97)07324-8
Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC (1997) Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother 40:135–136. https://doi.org/10.1093/jac/40.1.135
Maor Y, Hagin M, Belausov N, Keller N, Ben-David D, Rahav G (2009) Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J Infect Dis 199:619–624. https://doi.org/10.1086/596629
Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP (2001) A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47:399–403. https://doi.org/10.1086/596629
Matsuo M, Cui L, Kim J, Hiramatsu K (2013) Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains derived from hVISA clinical strain Mu3. Antimicrob Agents Chemother 57:5843e53. https://doi.org/10.1128/AAC.00425-13
Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC (2012) Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 56:5845e51. https://doi.org/10.1128/AAC.01139-12
The Clinical and Laboratory Standards Institute (2022) M100-Performance Standards for Antimicrobial Susceptibility Testing, 32nd edn. Malvern, Pennsylvania
Di Gregorio S, Haim MS, Famiglietti ÁMR, Di Conza J, Mollerach M (2023) Comparative genomics identifies novel genetic changes associated with oxacillin, vancomycin and daptomycin susceptibility in ST100 methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 12:372. https://doi.org/10.3390/antibiotics12020372
Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899. https://doi.org/10.1111/j.1600-0463.2007.apm_630.x
Stepanović S, Djukić N, Djordjević V, Djukić S (2003) Influence of the incubation atmosphere on the production of biofilm by staphylococci. Clin Microbiol Infect 9:955–958. https://doi.org/10.1046/j.1469-0691.2003.00676.x
Di Gregorio S, Perazzi B, Ordoñez AM, De Gregorio S, Foccoli M, Lasala MB et al (2015) Clinical, microbiological, and genetic characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a teaching hospital. Microb Drug Resist 21:25–34. https://doi.org/10.1089/mdr.2014.0190
Satola SW, Farley MM, Anderson KF, Patel JB (2011) Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with the population analysis profile method as the reference method. J Clin Microbiol 49:177–183. https://doi.org/10.1128/JCM.01128-10
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. https://doi.org/10.1093/nar/gkh063
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S et al (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357. https://doi.org/10.1093/nar/gkj102
Galperin MY, Makarova KS, Wolf YI, Koonin EV (2015) Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. https://doi.org/10.1093/nar/gku1223
Chen L, Xiong Z, Sun L, Yang J, Jin Q (2012) VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40:D641–D645. https://doi.org/10.1093/nar/gkr989
Liu B, Pop M (2009) ARDB-antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447. https://doi.org/10.1093/nar/gkn656
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12. https://doi.org/10.1186/gb-2004-5-2-r12
Castro BE, Rios R, Carvajal LP, Vargas ML, Cala MP, León L et al (2022) Multiomics characterization of methicillin-resistant Staphylococcus aureus (MRSA) isolates with heterogeneous intermediate resistance to vancomycin (hVISA) in Latin America. J Antimicrob Chemother. 78:122–132. https://doi.org/10.1093/jac/dkac363
Lamichhane-Khadka R, Dulal S, Cuaron JA, Pfeltz R, Gupta SK, Wilkinson BJ, Gustafson JE (2021) Apt (adenine phosphoribosyltransferase) mutation in laboratory-selected vancomycin-intermediate Staphylococcus aureus. Antibiotics (Basel) 10:583. https://doi.org/10.3390/antibiotics10050583
Gostev V, Kalinogorskaya O, Sopova J, Sulian O, Chulkova P, Velizhanina M et al (2023) Adaptive laboratory evolution of Staphylococcus aureus resistance to vancomycin and daptomycin: mutation patterns and cross-resistance. Antibiotics (Basel) 12:928. https://doi.org/10.3390/antibiotics12050928
Lecaillon E, Gueudet P, Wooton M, Walsh TR, Macgowan AP, Jones ME (2002) Endemic heteroresistant glycopeptide intermediate Staphylococcus aureus (hGISA) comprising unrelated clonal types and not associated with vancomycin therapy. Pathol Biol (Paris) 50:525–529. https://doi.org/10.1016/s0369-8114(02)00345-0
Yamakawa J, Aminaka M, Okuzumi K, Kobayashi H, Katayama Y, Kondo S et al (2012) Heterogeneously vancomycin-intermediate Staphylococcus aureus (hVISA) emerged before the clinical introduction of vancomycin in Japan: a retrospective study. J Infect Chemother 18:406–409. https://doi.org/10.1007/s10156-011-0330-2
Gaillard T, Dupieux-Chabert C, Butin M, Dumitrescu O, Naceur O, Bouveyron C et al (2022) Heterogeneous vancomycin resistance in Staphylococcus aureus does not predict development of vancomycin resistance upon vancomycin pressure. J Antimicrob Chemother 77:1032–1035. https://doi.org/10.1093/jac/dkab488
Turner J, Howe RA, Wootton M, Bowker KE, Holt HA, Salisbury V et al (2001) The activity of vancomycin against heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus explored using an in vitro pharmacokinetic model. J Antimicrob Chemother 48:727–730. https://doi.org/10.1093/jac/48.5.727
Khatib R, Sharma M, Johnson LB, Riederer K, Shemes S, Szpunar S (2015) Decreasing prevalence of isolates with vancomycin heteroresistance and vancomycin minimum inhibitory concentrations ≥2 mg/L in methicillin-resistant Staphylococcus aureus over 11 years: potential impact of vancomycin treatment guidelines. Diagn Microbiol Infect Dis 82:245–248. https://doi.org/10.1016/j.diagmicrobio.2015.03.014
Cheng X, Ma J, Su J (2022) An overview of analytical methodologies for determination of vancomycin in human plasma. Molecules 27:7319. https://doi.org/10.3390/molecules27217319
M S, Mulki SS, Shenoy S, Dhanashree B, M C, Bhat G (2023) Heterogeneous vancomycin intermediate Staphylococcus aureus infections in diabetic and non-diabetic patients – a hospital-based comparative study. Infect Drug Resist 16:9–17. https://doi.org/10.2147/IDR.S393415
Howden BP, Peleg AY, Stinear TP (2014) The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol 21:575–582. https://doi.org/10.1016/j.meegid.2013.03.047
Liang J, Hu Y, Fu M, Li N, Wang F, Yu X, Ji B (2023) Resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus and heterogeneous vancomycin-intermediate Staphylococcus aureus. Infect Drug Resist 16:379–388. https://doi.org/10.2147/IDR.S392908
Yang X, Qian S, Yao K, Wang L, Liu Y, Dong F et al (2017) Multiresistant ST59-SCCmec IV-t437 clone with strong biofilm-forming capacity was identified predominantly in MRSA isolated from Chinese children. BMC Infect Dis 17:733. https://doi.org/10.1186/s12879-017-2833-7
Chen X, Sun K, Luo Q, Duan Y, Chen F (2019) Emergence and spread of pvl-positive genotypic CA-MRSA ST59 with increased adhesion capacity from wounds in hospitals. J Infect 79:612–625. https://doi.org/10.1016/j.jinf.2019.10.005
Manasherob R, Mooney JA, Lowenberg DW, Bollyky PL, Amanatullah DF (2021) Tolerant small-colony variants form prior to resistance within a Staphylococcus aureus biofilm based on antibiotic selective pressure. Clin Orthop Relat Res 479:1471–1481. https://doi.org/10.1097/CORR.0000000000001740
Faúndez G, Troncoso M, Navarrete P, Figueroa G (2004) Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Campylobacter jejuni. BMC Microbiol 4:19. https://doi.org/10.1186/1471-2180-4-19
Yusof A, Engelhardt A, Karlsson A, Bylund L, Vidh P, Mills K et al (2008) Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J Clin Microbiol 46:3042–3047. https://doi.org/10.1128/JCM.00265-08
Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, Horikawa Y, Hiramatsu K (2006) Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:428–438. https://doi.org/10.1128/AAC.50.2.428-438.2006
Cui L, Tominaga E, Neoh HM, Hiramatsu K (2006) Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:1079–1082. https://doi.org/10.1128/AAC.50.3.1079-1082.2006
Kuroda M, Sekizuka T, Matsui H, Ohsuga J, Ohshima T, Hanaki H (2019) IS256-mediated overexpression of the WalKR two-component system regulon contributes to reduced vancomycin susceptibility in a Staphylococcus aureus Clinical Isolate. Front Microbiol 10:1882. https://doi.org/10.3389/fmicb.2019.01882
Di Gregorio S, Fernandez S, Perazzi B, Bello N, Famiglietti A, Mollerach M (2016) Increase in IS256 transposition in invasive vancomycin heteroresistant Staphylococcus aureus isolate belonging to ST100 and its derived VISA mutants. Infect Genet Evol 43:197–202. https://doi.org/10.1016/j.meegid.2016.05.001
Komatsuzawa H, Ohta K, Sugai M, Fujiwara T, Glanzmann P, Berger-BächiB SH (2000) Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J Antimicrob Chemother 45:421–431. https://doi.org/10.1093/jac/45.4.421
Thitiananpakorn K, Aiba Y, Tan XE, Watanabe S, Kiga K, Sato’o Y et al (2020) Association of mprF mutations with cross-resistance to daptomycin and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA). Sci Rep 10:16107. https://doi.org/10.1038/s41598-020-73108-x
Cameron DR, Ward DV, Kostoulias X, Howden BP, Moellering RC Jr, Eliopoulos GM, Peleg AY (2012) Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis 205:1677–1687. https://doi.org/10.1093/infdis/jis252
Hu Q, Peng H, Rao X (2016) Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol 7:1601. https://doi.org/10.3389/fmicb.2016.01601
Acknowledgements
The authors thank Dr. Yuanyuan Dai from the Provincial Hospital of Anhui Medical University in Hefei, China, for providing the control strains Mu3 and Mu50.
Funding
This study was financially supported by the National Key Research and Development Program of China (2021YFC2301004) and the National Key Inspection Specialty Construction Projects.
Author information
Authors and Affiliations
Contributions
Conceptualization: X.C., L.M., and J.S.; methodology: X.C. and Y.W.; software: X.C. and J.M.; validation: X.C. and W.S.; formal analysis: X.C. and Y.W.; investigation: X.C. and L.M.; resources: W.S.; data curation: X.C.; writing–original draft preparation: X.C.; writing, review, and editing: L.M. and J.S.; visualization: X.C.; supervision: J.S.; project administration: L.M. and J.S.; funding acquisition: J.S.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Ilana Camargo
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, X., Wang, Y., Ma, J. et al. Resistance phenotype and genetic features of a heterogeneous vancomycin intermediate–resistant Staphylococcus aureus strain from an immunocompromised patient. Braz J Microbiol 55, 323–332 (2024). https://doi.org/10.1007/s42770-023-01192-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-01192-y