Abstract
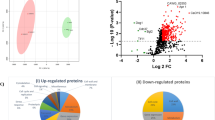
Chlamydospore though considered as a unique and rare morphological form of Candida albicans, regulation and significance of chlamydosporulation is not very clear. SWATH-MS analysis of chlamydosporulation specific proteins revealed that 319 (137-Up regulated and 182-Down regulated) proteins expressed differentially. Functional annotation showed significant modulations in proteins involved in cellular architecture (30), carbohydrate (29), amino acid (17), fatty acid (3), Nucleic acid (14), vitamins (1) metabolism as well as signaling (6), stress response (26), transport (cytoplasmic-21, mitochondrial-6 and nuclear-1), gene expression (transcription-12, RNA processing-6, translation-53, PTM-18), proteolysis (15) etc. Enhanced mannan, β1, 3-glucan and chitin contribute in thickening of cell wall while Hyr1 (218-fold) and Als3 (38.16-fold) dominates the cell surface chemistry of chlamydospores. In addition to ergosterol, enhanced sphingolipids, phospholipids and fatty acids make chlamydospore membrane more sturdy and rigid. Up-regulation of maltase (64-fold) followed by enhanced glycolysis and tricarboxylic acid cycle under nutrient-limiting condition is indicative of chlamydosporulation. Glyoxylate and fermentative pathway reported to facilitate survival of C. albicans under glucose limiting and microaerophilic condition was up-regulated. Enhanced biosynthesis of glutathione, trehalose homeostasis, and inhibition of NAD+ generation ,etc., potentiate oxidative, osmotic and nitrosative stress tolerance. Up regulation of Rsr1 (8.83-fold) and down regulation of Bcy1 (4.20-fold), Tfs1 (negative regulator of RAS) indicates cAMP-PKA pathway activates chlamydosporulation through Efg1 (a morphogenic regulator) in our study. In general, morpho-physiological modulations in C. albicans is a result of different sets of transcriptional programs that facilitate survival under nutrient and oxygen limiting condition.






Similar content being viewed by others
References
Aimanianda V, Clavaud C, Simenel C, Fontaine T, Delepierre M, Latgé JP (2009) Cell wall β-(1,6)-glucan of Saccharomyces cerevisiae structural characterization and in situ synthesis. J Biol Chem 284:13401–13412
Arthington-Skaggs BA, Jradi H, Desai T, Morrison CJ (1999) Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J Clin Microbiol 37:3332–3337
Barelle CJ, Priest CL, MacCallum DM, Gow NA, Odds FC, Brown AJ (2006) Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microb 8:961–971. https://doi.org/10.1111/j.1462-5822.2005.00676.x
Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA (2003) NAD+ -dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mole Cell Biol 23:7044–7054. https://doi.org/10.1128/MCB.23.19.7044-7054.2003
Berman J, Sudbery PE (2002) Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet 3:918–932. https://doi.org/10.1038/nrg948
Biswas S, Van Dijck P, Datta A (2007) Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev 71:348–376. https://doi.org/10.1128/MMBR.00009-06
Bottcher B, Pöllath C, Staib P, Hube B, Brunke S (2016) Candida species rewired hyphae developmental programs for chlamydospore formation. Front Microb 7:1697. https://doi.org/10.3389/fmicb.2016.01697
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Brown AJ, Budge S, Kaloriti D, Tillmann A, Jacobsen MD, Yin Z, Ene IV, Bohovych I, Sandai D, Kastora S, Potrykus J (2014) Stress adaptation in a pathogenic fungus. J Ex Biol 217:144–155. https://doi.org/10.1242/jeb.088930
Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP (2006) Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog 2:e21. https://doi.org/10.1371/journal.ppat.0020021
Cabezon V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C (2009) Analysis of Candida albicans plasma membrane proteome. Proteomics 9:4770–4786. https://doi.org/10.1002/pmic.200800988
Cabrera M, Arlt H, Epp N, Lachmann J, Griffith J, Perz A, Reggiori F, Ungermann C (2013) Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J Biol Chem 288:5166–5175. https://doi.org/10.1074/jbc.M112.431536
Cao YY, Cao YB, Xu Z, Ying K, Li Y, Xie Y, Jiang YY (2005) cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother 49:584–589. https://doi.org/10.1128/AAC.49.2.584-589.2005
CDC Report (2013) Antibiotic resistance threats in the United States
Cederquist GY, Luchniak A, Tischfield MA, Peeva M, Song Y, Menezes MP, Chan WM, Andrews C, Chew S, Jamieson RV, Gomes L (2012) An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet 21:5484–5499. https://doi.org/10.1093/hmg/dds393
CGD (2010) Description lines for gene products based on orthologs and predicted Gene Ontology (GO)
Chauhan NM, Raut JS, Karuppayil SM (2011) A morphogenetic regulatory role for ethyl alcohol in Candida albicans. Mycoses 54:6. https://doi.org/10.1111/j.1439-0507.2010.02002.x
Chautard H, Jacquet M, Schoentgen F, Bureaud N, Bénédetti H (2004) Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae. Eukaryot Cell 3:459–470. https://doi.org/10.1128/EC.3.2.459-470.2004
Chen YL, Kauffman S, Reynolds TB (2008) Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect Immun 76:2793–2801. https://doi.org/10.1128/IAI.01514-07
Citiulo F, Moran GP, Coleman DC, Sullivan DJ (2009) Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Res 9:1051–1060. https://doi.org/10.1111/j.1567-1364.2009.00533.x
Collins BC, Gillet LC, Rosenberger G, Röst HL, Vichalkovski A, Gstaiger M, Aebersold R (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods 10:1246–1253. https://doi.org/10.1038/nmeth.2703
Collinson EJ, Wheeler GL, Garrido EO, Avery AM, Avery SV, Grant CM (2002) The yeast glutaredoxins are active as glutathione peroxidases. J Biol Chem 277:16712–16717. https://doi.org/10.1128/AAC.48.8.3064-3079.2004
Cottier F, Raymond M, Kurzai O, Bolstad M, Leewattanapasuk W, Jiménez-López C, Lorenz MC, Sanglard D, Váchová L, Pavelka N, Palková Z (2012) The bZIP transcription factor Rca1p is a central regulator of a novel CO2 sensing pathway in yeast. PLoS Pathog 8:e1002485. https://doi.org/10.1371/journal.ppat.1002485
Curwin AJ, von Blume J, Malhotra V (2012) Cofilin-mediated sorting and export of specific cargo from the Golgi apparatus in yeast. Mol Biol Cell 23:2327–2338. https://doi.org/10.1091/mbc.e11-09-0826
Cutler JE (1991) Putative virulence factors of Candida albicans. Annu Rev Microbiol 45:187–218
Dong K, Addinall SG, Lydall D, Rutherford JC (2013) The yeast copper response is regulated by DNA damage. Mol Cell Biol 33:4041–4050. https://doi.org/10.1128/MCB.00116-13
Douglas LM, Alvarez FJ, McCreary C, Konopka JB (2005) Septin function in yeast model systems and pathogenic fungi. Eukaryot Cell 4:1503–1512. https://doi.org/10.1128/EC.4.9.1503-1512.2005
Eisman B, Alonso-Monge R, Roman E, Arana D, Nombela C, Pla J (2006) The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5:347–358. https://doi.org/10.1128/EC.5.2.347-358.2006
Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NA, Brown AJ (2012) Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14:1319–1335. https://doi.org/10.1111/j.1462-5822.2012.01813.x
Ener B, Douglas LJ (1992) Correlation between cell-surface hydrophobicity of Candida albicans and adhesion to buccal epithelial cells. FEMS Microbiol Lett 99(1):37–42. https://doi.org/10.1111/j.1574-6968.1992.tb05538.x
Ernst JF (2000) Transcription factors in Candida albicans–environmental control of morphogenesis. Microbiology 146:1763–1774. https://doi.org/10.1099/00221287-146-8-1763
Fu MS, De Sordi L, Mühlschlegel FA (2012) Functional characterization of the small heat shock protein Hsp12p from Candida albicans. PLoS ONE 7:e42894. https://doi.org/10.1371/journal.pone.0042894
García-Sánchez S, Mavor AL, Russell CL, Argimon S, Dennison P, Enjalbert B, Brown AJ (2005) Global roles of Ssn6 in Tup1-and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell 16:2913–2925. https://doi.org/10.1091/mbc.e05-01-0071
Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteom 11:O111–016717. https://doi.org/10.1074/mcp.O111.016717
Giosa D, Felice MR, Lawrence TJ, Gulati M, Scordino F, Giuffrè L, Lo Passo C, D’Alessandro E, Criseo G, Ardell DH, Hernday AD (2017) Whole RNA-sequencing and transcriptome assembly of Candida albicans and Candida africana under chlamydospore-inducing conditions. Genome Biol Evol 9:1971–1977. https://doi.org/10.1093/gbe/evx143
Gleason JE, Li CX, Odeh HM, Culotta VC (2014) Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans. J Biol Inorg Chem 19:595–603. https://doi.org/10.1007/s00775-013-1045-x
Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL (2004) RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 150:267–275. https://doi.org/10.1099/mic.0.26699-0
Guthrie C, Fink GR (eds) (2002) Guide to yeast genetics and molecular and cell biology: part C. Gulf Professional Publishing, Houston
Haar VDT (2007) Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE 2:e1078. https://doi.org/10.1371/journal.pone.0001078
Hall RA (2015) Dressed to impress: impact of environmental adaptation on the Candida albicans cell wall. Mol Microbiol 97:7–17. https://doi.org/10.1111/mmi.13020
Han TL, Cannon RD, Villas-Bôas SG (2011) The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet Biol 48:747–763. https://doi.org/10.1016/j.fgb.2011.04.002
Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M (2004) Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell 15:4490–4499. https://doi.org/10.1091/mbc.e04-02-0144
Hazel JR, Williams EE (1990) The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog Lipid Res 29:167–227. https://doi.org/10.1016/0163-7827(90)90002-3
Hazen KC (1990) Cell surface hydrophobicity of medically important fungi, especially Candida species. In Microbial Cell Surface Hydrophobicity. ASM Press, Washington, DC, USA, pp 249–295
Hazen KC, Hazen BW (1987) A polystyrene microsphere assay for detecting surface hydrophobicity variations within Candida albicans populations. J Microbiol Methods 6:289–299. https://doi.org/10.1016/0167-7012(87)90066-2
Hazen KC, Plotkin BJ, Klimas DM (1986) Influence of growth conditions on cell surface hydrophobicity of Candida albicans and Candida glabrata. Infect Immun 54:269–271
He XY, Meurman JH, Kari K, Rautemaa R, Samaranayake LP (2006) In vitro adhesion of Candida species to denture base materials. Mycoses 49:80–84
Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, de Koster CG, de Koning LJ, Klis FM (2011) Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157:2297–2307. https://doi.org/10.1099/mic.0.049395-0
Holmes AR, Shepherd MG (1987) Proline-induced germ-tube formation in Candida albicans: role of proline uptake and nitrogen metabolism. Microbiology 133:3219–3228. https://doi.org/10.1099/00221287-133-11-3219
Ingle S, Kodgire S, Shiradhone A, Patil R, Zore G (2017) Chlamydospore specific proteins of Candida albicans. Data 2:26. https://doi.org/10.3390/data2030026
Jamai L, Ettayebi K, El Yamani J, Ettayebi M (2007) Production of ethanol from starch by free and immobilized Candida tropicalis in the presence of α-amylase. Bioresour Technol 98:2765–2770. https://doi.org/10.1016/j.biortech.2006.09.057
Jansons VK, Nickerson WJ (1970) Chemical composition of chlamydospores of Candida albicans. J Bacteriol 104:922–932
Johnston DA, Tapia AL, Eberle KE, Palmer GE (2013) Three prevacuolar compartment Rab GTPases impact Candida albicans hyphal growth. Eukaryot Cell 12:1039–1050. https://doi.org/10.1128/EC.00359-12
Kamthan M, Mukhopadhyay G, Chakraborty N, Chakraborty S, Datta A (2012) Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet Biol 49:369–378. https://doi.org/10.1016/j.fgb.2012.02.006
Karababa M, Coste AT, Rognon B, Bille J, Sanglard D (2004) Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother 48:3064–3079. https://doi.org/10.1128/AAC.48.8.3064-3079.2004
Kato M, Lin SJ (2014) YCL047C/POF1 is a novel nicotinamide mononucleotide adenylyltransferase (NMNAT) in Saccharomyces cerevisiae. J Biol Chem 289:15577–15587. https://doi.org/10.1074/jbc.M114.558643
Kelly JP, Funigiello F (1959) Candida albicans: a study of media designed to promote chlamydospore production. J Lab C Med 53:807–809
Kim D, Shin WS, Lee KH, Kim K, Young Park J, Koh CM (2002) Rapid differentiation of Candida albicans from other Candida species using its unique germ tube formation at 39 C. Yeast 19(11):957–962. https://doi.org/10.1002/yea.891
Klis FM, Sosinska GJ, De Groot PW, Brul S (2009) Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res 9:1013–1028. https://doi.org/10.1111/j.1567-1364.2009.00541.x
Klotz SA (1990) Role of hydrophobic interactions in microbial adhesion to plastic used in medical devices. Micro Cell Surf Hydrophobicity 12:107–136
Klotz SA, Penn RL (1987) Multiple mechanisms may contribute to the adherence of Candida yeasts to living cells. Curr Microbiol 16(3):119–122. https://doi.org/10.1007/bf01568389
Klotz SA, Drutz DJ, Zajic JE (1985) Factors governing adherence of Candida species to plastic surfaces. Infect Immun 50(1):97–101
Kullberg BJ, Arendrup MC (2015) Invasive candidiasis. N Engl J Med 373:1445–1456. https://doi.org/10.1056/NEJMra1315399
Lee KK, MacCallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NAR, Munro CA (2012) Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother 56:208–217. https://doi.org/10.1128/AAC.00683-11
Lim CY, Rosli R, Seow HF, Chong PP (2012) Candida and invasive candidiasis: back to basics. Eur J Clin Microbiol Infect Dis 31:21–31. https://doi.org/10.1007/s10096-011-1273-3
Lin MC, Galletta BJ, Sept D, Cooper JA (2010) Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci 123:1329–1342. https://doi.org/10.1242/jcs.065698
Liu L, Zeng M, Hausladen A, Heitman J, Stamler JS (2000) Protection from nitrosative stress by yeast flavohemoglobin. Proc Natl Acad Sci 97:4672–4676. https://doi.org/10.1073/pnas.090083597
Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Smith RD (2006) Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteom 5:2167–2174. https://doi.org/10.1074/mcp.T600039-MCP200
Liu MS, Li HC, Lai YM, Lo HF, Chen LF (2013) Proteomics and transcriptomics of broccoli subjected to exogenously supplied and transgenic senescence-induced cytokinin for amelioration of postharvest yellowing. J Proteom 93:133–144. https://doi.org/10.1016/j.jprot.2013.05.014
Liu Q, Han Q, Wang N, Yao G, Zeng G, Wang Y, Huang Z, Sang J, Wang Y (2016) Tpd3-Pph21 phosphatase plays a direct role in Sep7 dephosphorylation in Candida albicans. Mol Microbiol 101:109–121. https://doi.org/10.1111/mmi.13376
Lorenz MC, Bender JA, Fink GR (2004) Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. https://doi.org/10.1128/EC.3.5.1076-1087.2004
Martin SW, Douglas LM, Konopka JB (2005) Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot Cell 4:1191–1202. https://doi.org/10.1128/EC.4.7.1191-1202.2005
Mayer FL, Wilson D, Jacobsen ID, Miramón P, Slesiona S, Bohovych IM, Brown AJ, Hube B (2012) Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS ONE 7:e38584. https://doi.org/10.1371/journal.pone.0038584
Mc Manus BA, Coleman DC (2014) Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect Genet Evol 21:166–178. https://doi.org/10.1016/j.meegid.2013.11.008
Michán C, Pueyo C (2009) Growth phase-dependent variations in transcript profiles for thioredoxin-and glutathione-dependent redox systems followed by budding and hyphal Candida albicans cultures. FEMS Yeast Res 9:1078–1090. https://doi.org/10.1111/j.1567-1364.2009.00558.x
Miller SE, Spurlock BO, Michaels GE (1974) Electron microscopy of young Candida albicans chlamydospores. J Bacteriol 119:992–999
Mun MS, Yap T, Alnuaimi AD, Adams GG, McCullough MJ (2016) Oral candidal carriage in asymptomatic patients. Aust Dent J 61:190–195. https://doi.org/10.1111/adj.12335
Navarathna DH, Pathirana RU, Lionakis MS, Nickerson KW, Roberts DD (2016) Candida albicans ISW2 regulates chlamydospore suspensor cell formation and virulence in vivo in a mouse model of disseminated candidiasis. PLoS ONE 11:e0164449. https://doi.org/10.1371/journal.pone.0164449
Nett JE, Lepak AJ, Marchillo K, Andes DR (2009) Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200:307–313. https://doi.org/10.1086/599838
Neville BA, d’Enfert C, Bougnoux ME (2015) Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fov081
Nobile CJ, Bruno VM, Richard ML, Davis DA, Mitchell AP (2003) Genetic control of chlamydospore formation in Candida albicans. Microbiology 149:3629–3637. https://doi.org/10.1099/mic.0.26640-0
Odds FC (1988) Candida and Candidosis. Baillière Tindall, London
Odds FC, Bernaerts RIA (1994) CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol 32(8):1923–1929
Palige K, Linde J, Martin R, Böttcher B, Citiulo F, Sullivan DJ, Johann W, Claudia S, Staib C, Steffen R, Bernhard H, Morschhäuser J, Peter Staib J (2013) Global transcriptome sequencing identifies chlamydospore specific markers in Candida albicans and Candida dubliniensis. PLoS ONE 8(4):e61940. https://doi.org/10.1371/journal.pone.0061940
Panagoda GJ, Ellepola ANB, Samaranayake LP (2001) Adhesion of Candida parapsilosis to epithelial and acrylic surfaces correlates with cell surface hydrophobicity. Mycoses 44(1–2):29–35
Pereira MD, Eleutherio EC, Panek AD (2001) Acquisition of tolerance against oxidative damage in Saccharomyces cerevisiae. BMC Microbiol 1:11. https://doi.org/10.1186/1471-2180-1-11
Richard M, de Groot P, Courtin O, Poulain D, Klis F, Gaillardin C (2002) GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans. Microbiology 148:2125–2133. https://doi.org/10.1099/00221287-148-7-2125
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9(1):29–33
Ruhnke M (2006) Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Curr Drug Targets 7:495–504. https://doi.org/10.2174/138945006776359421
Samaranayake YH, Samaranayake LP, Yau JYY, Ellepola ANB, Anil S, Yeung KWS (2003) Adhesion and cell-surface-hydrophobicity of sequentially isolated genetic isotypes of Candida albicans in an HIV-infected Southern Chinese cohort. Mycoses 46(9–10):375–383. https://doi.org/10.1046/j.0933-7407.2003.00919.x
Sarkar P, Florczyk M, McDonough K, Nag D (2002) SSP2, a sporulation-specific gene necessary for outer spore wall assembly in the yeast Saccharomyces cerevisiae. Mol Genet Genom 267:348–358. https://doi.org/10.1007/s00438-002-0666-5
Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, Sutphin GL (2013) Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell 12:1050–1061. https://doi.org/10.1111/acel.12130
Seneviratne CJ, Wang Y, Jin L, Abiko Y, Samaranayake LP (2008) Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 14:2936–2947. https://doi.org/10.1002/pmic.200701097
Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J (2004) A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15:4179–4190. https://doi.org/10.1091/mbc.e04-03-0181
Smits GJ, van den Ende H, Klis FM (2001) Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781–794. https://doi.org/10.1099/00221287-147-4-781
Sonneborn A, Bockmühl DP, Ernst JF (1999) Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect Immun 67:5514–5517
Sosinska GJ (2012) Adaptations in the wall proteome of the clinical fungus Candida albicans in response to infection-related environmental conditions, Ph.D. Thesis, University of Amsterdam, Netherland
Staib P, Morschhäuser J (2005) Liquid growth conditions for abundant chlamydospore formation in Candida dubliniensis. Mycoses 48(1):50–54. https://doi.org/10.1111/j.1439-0507.2004.01085.x
Staib P, Morschhäuser J (2007) Chlamydospore formation in Candida albicans and Candida dubliniensis–an enigmatic developmental programme. Mycoses 50:1–12. https://doi.org/10.1111/j.1439-0507.2006.01308.x
Tyc KM, Kühn C, Wilson D, Klipp E (2014) Assessing the advantage of morphological changes in Candida albicans: a game theoretical study. Front Microbiol 5:41. https://doi.org/10.3389/fmicb.2014.00041
Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11:3425–3439. https://doi.org/10.1091/mbc.11.10.3425
Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P (2006) Eisosomes mark static sites of endocytosis. Nature 439:998. https://doi.org/10.1038/nature04472
Williams SM (2011) Investigating the role of Nrg1p and Tup1p during Candida albicans chlamydospore formation. McNair Scholars J 15:11
Yadav AK, Desai PR, Rai MN, Kaur R, Ganesan K, Bachhawat AK (2011) Glutathione biosynthesis in the yeast pathogens Candida glabrata and Candida albicans: essential in C. glabrata, and essential for virulence in C. albicans. Microbiology 157:484–495. https://doi.org/10.1099/mic.0.045054-0
Yoda K, Kawada T, Kaibara C, Fujie A, Abe M, Hashimoto H, Shimizu J, Tomishige N, Noda Y, Yamasaki M (2000) Defect in cell wall integrity of the yeast Saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene VIG9. Biosci Biotechnol Biochem 64:1937–1941. https://doi.org/10.1271/bbb.64.1937
Young ME, Karpova TS, Brügger B, Moschenross DM, Wang GK, Schneiter R, Cooper JA (2002) The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol Cell Biol 22:927–934. https://doi.org/10.1128/MCB.22.3.927-934.2002
Acknowledgements
Authors are thankful to Prof. Udhav V. Bhosle, Honorable Vice Chancellor, SRTM University, Nanded (MS) India for his encouragement and incessant support. Authors are also thankful to SERB, India for financial support under SERB FAST Track Scheme for Young Scientists to GBZ. GBZ acknowledge geneorous financial support of UGC under UGC-SAP-DRS II and DST under DST-FIST I to the School of Life Sciences, SRTM University, Nanded.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ingle, S., Kazi, R., Patil, R. et al. Proteome analysis of Candida albicans cells undergoing chlamydosporulation. J Proteins Proteom 10, 269–290 (2019). https://doi.org/10.1007/s42485-019-00024-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-019-00024-8