Abstract
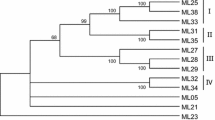
Management of chickpea wilt incited by Fusarium oxysporum f. sp. ciceris has been done primarily through development and use of resistant cultivars as part of an integrated management approach as the resistance genes are a major factor behind protecting plants from various pathogens. Thus, isolation and characterization of the RGAs of chickpea provide a critical foundation for deciphering host-pathogen interactions and development of novel methods to manage pathogens of crop plant. Eleven RGAs were identified in resistant variety (WR 315) of chickpea by degenerate PCR amplification followed by sequencing, cloning and database search. The restriction pattern generated using enzyme Hinf1 as well as analysis of phylogenetic tree of derived amino acid sequences classified chickpea RGA sequences into four classes of RGAs. Amino acid sequence alignment of these clones clearly showed the presence of consensus motifs namely, P-loop and Non-TIR linked sequences. The finding further supports that these consensus motifs are extensively present in dicot plant species. Three conserved motifs within the aligned sequences were also identified using MEME analysis. These conserve motifs exhibited the high level of similarity with NB-ARC domain as shown by pfam protein motif analyse. Quantitative real time-PCR analysis clearly revealed that both SA and JA induce the expression of RGA genes, however, the expression was higher in the plants treated with SA as compared to the control or JA treated plants. The differentially expressed signaling molecules suggesting that SA and JS as stimuli mainly involved in responding defence and activating signaling pathways.







Similar content being viewed by others
References
Aarts N, Metz M, Holub E, Stakawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene mediated signaling pathways in Arabidopsis. Proc Nalt Acad Sci USA 95:10306–10311
Angela F, Angelica MJ, Laurent T, Ralph P, Lan BD (2008) Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol 35:1255–1266
An C, Mou Z (2011) Salicylic acid and its function in plant immunity. J Integr Plant Biol 53:412–428
Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29
Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6:170
Cheng Y, Xiaoyu L, Haiyang J, Wei M, Weiyun M, Toshihiko Y, Ming Z (2012) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J 279:2431–2443
Dubey SC (2016) Race profiling, genetic diversity, diagnostics and management of Fusarium oxysporum f. sp. ciceris causing wilt in chickpea. Indian Phytopathol 69:210–217
Dubey SC, Singh SR (2008) Virulence analysis and oligonucleotide fingerprinting to detect diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165:389–406
Dubey N, Singh K (2018) Role of NBS-LRR proteins in plant defense. In: Singh A, Singh I (eds) Molecular aspects of plant-pathogen interaction. Springer, Cham, pp 115–138
Dubey SC, Tripathi A, Tak R (2018) Expression of defense-related genes in mung bean varieties in response to Trichoderma virens alone and in the presence of Rhizoctonia solani infection. 3 Biotech 8:432
Fitzgerald TL, McQualter RB (2014) The quantitative real-time polymerase chain reaction for the analysis of plant gene expression. Cereal Genom 1099:97–115
Flor HH (1956) Complementary genetic system in flax and flax rust. Adv Genet 8:29–54
Guttman DS, McHardy AC, Schulze-Lefert P (2014) Microbial genome enabled insights into plant-microorganism interactions. Nat Rev Genet 15:797–813
Hall T (2011) BioEdit: an important software for molecular biology. GERF Bulletin of Biosci 2:60–61
Haware MP, Nene YL (1980) Influence of wilt at different stages on the yield loss in chickpea. Trop Grain Legume Bull 19:38–40
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kozjak P, Jakse J, Javornik B (2009) Isolation and sequence analysis of NBS-LRR disease resistance gene analogues from hop Humulus lupulus L. Plant Sci 176:775–782
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Lozano-Torres JL, Wilbers RHP, Gawronski P, Boshoven JC, Finkers Tomczak A, Cordewener JH et al (2012) Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc Natl Acad Sci 109:10119–10124
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Murray MG, Thompson (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Ooijen GV, Mayr G, Kasiem MMA, Albrecht M, Cornelissen BJC, Takken FLW (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59:1383–1397
Palomino C, Satovic Z, Cubero JI, Torres AM (2006) Identification and characterization NBS-LRR class resistance gene analogs in Fababean (Viciafaba L.) and chickpea (Cicer arietinum L.). Genome 49:1227–1237
Palomino C, Ferna´ndez-Romero MD, Rubio J, Torres A, Moreno MT, Milla´n T (2009) Integration of new CAPS and dCAPS-RGA markers into a composite chickpea genetic map and their association with disease resistance. Theor Appl Genet 118:671–682
Rachana KE, Naganeeswaran SA, Fayas TP, Thomas RJ, Rajesh MK (2016) Cloning, characterization and expression analysis of NBS-LRR-type resistance gene analogues (RGAs) in coconut. Acta Bot Croat 75:1–10
Rafael M, Jimenez D, Juan A, Navas C (2015) Fusarium wilt of chickpeas: biology, ecology and management. Crop Prot 73:16–27
Sharma M, Ghosh R, Tarafdar A, Rathore A, Chobe DR, Kumar A, Gaur PM, Samineni S, Gupta O, Singh NP, Saxena DR, Saifulla M, Pithia MS, Ghante PH, Mahalinga DM, Upadhyay JB, Harer PN (2019) Exploring the genetic cipher of Chickpea (Cicer arietinum L.) through identification and multi-environment validation of resistant sources against Fusarium wilt (Fusarium oxysporum f. sp. ciceris). Front Sustain Food Syst 3:78
Shirano Y, Kachroo P, Shah J, Klessig DF (2002) A gain-of-function mutation in an Arabidopsis toll interleukin1 receptor-nucleotide binding site-Leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14:3149–3162
Speulman E, Bouchez D, Holub EB, Beynon JL (1998) Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana. Plant J 14:467–474
Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD (1995) Molecular genetics of plant disease resistance. Science 268:661–667
Steinbrenner AD, Goritschnig S, Staskawicz BJ (2015) Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog 11:e1004665
Takken FLW, Goverse A (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol 15:375–384
Tameling WIL, Elzinga SD, Darmin PS, Vossen JH, Takken FLW, Haring MA et al (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14:2929–2939
Tan S, Wu S (2012) Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp Funct Genom 2012:418208
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleiotide-binding sites. Eur J Biochem 222:9–19
Wang Y, Rosen B, Scoffield J, Egnin M, Mortley D, Steiner S, Cook DR, He G (2010) Isolation and analysis of resistance gene homologues in sweet potato. Plant Breed 129:519–525
Wang Y, Song Z, Zhang W, Xu L, Su X, Mmbone E, LW L (2017) Identification and characterization of expressed TIR- and non-TIR-NBS-LRR resistance gene analogous sequences from radish (Raphanus sativus L.) de novo transcriptome. Sci Hortic 216:284–292
Wei H, Li W, Sun X, Zhu S, Zhu J (2013) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS One 8:e68435
Xiao SX, Brown EP, Brearley C, Turner JG (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15:33–45
Xiong QY, Wei LJ, Sen ZJ, Hong RM, Ping XL, Qing ZM (2008) Molecular cloning and characterisation of a non-TIR-NBS-LRR type disease resistance gene analogue from sugarcane. Sugar Tech 10:71–73
Yang DL, Yao J, Mei CS, Tong XH et al (2012) Plant hormone jasmonate priorities defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109:1192–1200
Zhou Z, Bar I, Sambasivam PT, Ford R (2019) Determination of the key resistance gene analogs involved in Ascochyta rabiei recognition in chickpea. Front Plant Sci 10:644
Acknowledgements
The authors gratefully acknowledge the financial support of the Indian Council of Agricultural Research (ICAR), New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Priyanka, K., Dubey, S.C. & Upadhyay, B.K. Cloning, characterization and expression analysis of resistant gene analogues for wilt resistant in chickpea. Indian Phytopathology 74, 649–658 (2021). https://doi.org/10.1007/s42360-021-00363-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-021-00363-x