Abstract
A multiplex reverse transcription polymerase chain reaction (multiplex RT-PCR) assay was developed to enable the simultaneous detection and differentiation of four viruses that infect passion fruit, citrus-associated rhabdovirus (CiaRV), East Asian Passiflora virus (EAPV), Passiflora latent virus (PLV), and Telosma mosaic virus (TeMV). The optimized parameters included the primer concentration, annealing temperature, extension time, and number of cycles. The established multiplex RT-PCR assay produced the corresponding products with sizes of 597 bp, 529 bp, 320 bp, and 235 bp, which were specific for CiaRV, EAPV, PLV, and TeMV, respectively. The fragments could be distinguished clearly by agarose gel electrophoresis. The detection limit of the assay was 100 pg of total RNA for CiaRV and EAPV, 10 pg of total RNA for PLV, and 1.0 ng of total RNA for TeMV. The multiplex RT-PCR assay was also tested using field samples, and the results showed that the developed assay could detect the viruses in single or multiple infections of passion fruit. The multiplex RT-PCR established here will be quite helpful for the diagnosis of passion fruit infected with various combinations of the four viruses in the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Passion fruit (Passiflora edulis Sims), originated in South America, has been widely cultivated in tropical and subtropical regions worldwide because of its high pharmacological and edible value (Thokchom and Mandal 2017; Kawakami et al. 2021). In China, passion fruit orchards are located mainly in the southern part of the country, in Guangxi, Fujiang, and Jinagxi provinces. However, the cultivation and production of passion fruit are severely affected by various pathogens, such as viruses, bacteria, and fungi. Among them, viral diseases are extremely serious on passion fruit and a yield-limiting factor for the crop. It has been documented that passion fruit is susceptible to infection by more than 30 different viruses (Baker et al. 2011). These include potyviruses, such as East Asian Passiflora virus (EAPV) (Iwai et al. 2006), Telosma mosaic virus (TeMV) (Chiemsombat et al. 2014), and passion fruit woodiness virus (PWV) (Sokhandan et al. 1997); carlaviruses, such as Passiflora latent virus (PLV) (Spiegel et al. 2007); begomoviruses, as passion fruit severe leaf distortion virus (Ferreira et al. 2010), Euphorbia leaf curl virus (ELCV), papaya leaf curl Guangdong virus (PaLCuGDV) (Cheng et al. 2014), and papaya leaf curl China virus (PaLCuCNV) (Huang et al. 2019); and tentative cytorhabdoviruses, such as citrus-associated rhabdovirus (CiaRV) (Zhang et al. 2020). Many of the known passion fruit viruses have been detected in China, such as TeMV, EAPV, CiaRV, and PLV. TeMV and EAPV are both considered causal agents of passion fruit woodiness disease, which is characterized by foliar rugosity, mosaic and distorted woody, and severely malformed fruits (Iwai et al. 2006; Chiemsombat et al. 2014), and have been detected in many passion fruit production areas in China (Xie et al. 2017, 2018; Chen et al. 2018). TeMV is especially pervasive in passion fruit plants in that country (Yu et al. 2021). CiaRV is a tentative cytorhabdovirus that can infect passion fruit, but the symptoms induced in the host are still not clear. Based on our previous investigation, the detection rate of CiaRV is also high, and CiaRV always coinfects with TeMV. PLV causes foliar and fruit mosaic symptoms on Passiflora spp. and has been reported to occur naturally in Germany, Australia, the USA, New Zealand, and Korea (Spiegel et al. 2007; Pares et al. 2007; Tang et al. 2008; Cho et al. 2021). We also detected PLV in local field passion fruit plants. Based on our previous survey, these four viruses were most commonly found in mixed infections on the local field passion fruit plants, especially the TeMV, EAPV, and CiaRV. A highly efficient and reliable detection method would be extremely valuable for these viral diseases’ management.
Currently, several diagnostic methods have been developed for the detection of TeMV and PLV, such as reverse transcription-polymerase chain reaction (RT-PCR), reverse transcription loop mediated isothermal amplification (RT-LAMP), and serological method (Spiegel et al. 2007; Chiemsombat et al. 2014; Fu et al. 2021). RT-PCR is the only available method for EAPV and CiaRV. However, these methods can only detect one virus in a single reaction. Multiplex RT-PCR, a variant of PCR, is a quick, reliable, and cost-effective method that has been used to successfully detect a variety of viruses simultaneously in a single assay. To date, multiplex RT-PCR has been used widely to diagnose infection in citrus, apples, cherry, peach, and papaya plants by related viruses (Osman et al. 2015; Hao et al. 2016; Noorani et al. 2013; Yu et al. 2013; Tuo et al. 2014).
However, the simultaneous detection of CiaRV, EAPV, PLV, and TeMV from infected passion fruit has not been reported. The main goal of this study was therefore to develop a sensitive and specific multiplex RT-PCR assay for the detection of four economically important viruses. The optimized multiplex RT-PCR assay was evaluated by detecting these viruses in field samples, and proven to be reliable and sensitive.
Materials and methods
Plants and viruses
A single passion fruit plant (Fig. 1) confirmed to be coinfected with all four viruses was used as the virus source to develop the multiplex RT-PCR assay. Total RNA was extracted from the leaves of passion fruit plants using RNA-easy isolation reagent (Vazyme Bio Inc., Nanjing, China) according to the manufacturer’s instructions. The integrity of the RNA was confirmed by the standard electrophoresis technique. Additionally, RNA quality and quantity were assessed using a NanoDrop 2000c spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA), after which the RNA was immediately stored at − 80 ℃ for further procedures.
Design of virus-specific primers
The primer PLVReh–RT–F/R was selected from published articles for simplex and multiplex RT-PCR (Jover-Gil et al. 2018; Spiegel et al. 2007). The respective virus-specific primers for CiaRV, EAPV, and TeMV RT-PCR assays were designed using Oligo 7.0 software. They were based on the conserved region of each virus’s coat protein (CP) or nucleocapsid protein (N) gene obtained by comparing the available genomic sequences of each virus in the NCBI GenBank (Table S1) to determine the RNA region with the least amount of sequence variability. The primer names, oligonucleotide sequences, expected size of amplicon, and references are given in Table 1. The specificity of the primers was verified using BLASTn from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). All primers were synthesized by GenScript Biotech Corporation (Nanjing, China). The multiplex RT-PCR products of the expected size were cloned into the pMD19-T cloning vector (TaKaRa), sequenced, and subjected to BLASTn search.
Simplex RT-PCR assays
The simplex RT-PCR reaction was conducted using a one-step RT-PCR kit (Vazyme Bio Inc., Nanjing, China) in a 20-μL reaction mixture containing 10 μL of 2 × one-step mix, 0.4 μL of each primer (10 pM), 1 μL of one-step Enzyme mix, 1 μL of template RNA, and 8.2 μL of RNase-free ddH2O. The amplification was carried out using a GeneAmp PCR System (Applied Biosystems, CA, USA) with the following parameters: a 30-min incubation step at 50 °C, followed by one cycle at 94 °C for 3 min, 35 cycles of denaturation at 94 °C for 20 s, annealing at 56 °C for 20 s, and extension at 72 °C for 50 s, followed by a final extension step of 72 °C for 7 min. The amplicons were visualized by 1.5% agarose gel electrophoresis. Each amplified viral target fragment was purified using a gel extraction kit (TaKaRa Bio Inc., Japan), cloned into a pMD-19 T vector (TaKaRa Bio Inc., Japan), and sequenced (Genewiz, China). The obtained nucleotide sequences were verified by BLASTn from NCBI.
Optimization of multiplex RT-PCR
The multiplex RT-PCR was optimized by sequentially testing the primer concentration, annealing temperature, extension time, and number of cycles using the plant RNA prepared from the virus source plant. To optimize the multiplex RT-PCR system, combinations of different concentrations of four primer pairs were tested. The concentrations of the four primers were tested from 0.15 to 0.30 pM. The annealing temperature was increased from 52 to 59 °C in increments of 1 °C. The number of cycles was set as 30, 35, and 40. The extension times tested were 45 s, 60 s, and 75 s. The multiplex RT-PCR amplifications were performed in a 20-μL reaction volume comprising 10 μL of 2 × one step mix, 0.15–0.30 pM EAPV–F/R primers, 0.15 pM of CiaRV–F/R, PLVReh–RT–F/R, or TeMV–F/R primers, 1 μL of one step enzyme mix, 1 μL template RNA, and RNase-free ddH2O to make up the volume to 20 μL. Multiplex RT-PCR amplification parameters were as follows: a 30-min incubation step at 50 °C, followed by one cycle at 94 °C for 3 min, (30–40) cycles of denaturation at 94 °C for 20 s, annealing at (52–59) °C for 20 s, and extension at 72 °C for (45–75) s, followed by a final extension step of 72 °C for 7 min. The amplicons were visualized by 1.5% agarose gel electrophoresis.
Detection limits of multiplex RT-PCR
The sensitivity of the optimized multiplex RT-PCR for all the four viruses was compared to those of the simplex RT-PCRs for CiaRV, EAPV, PLV, and TeMV, using tenfold plant total RNA prepared from the virus source plant (100 ng/μL, 10 ng/μL, 1.0 ng/μL, 100 pg/μL, 10 pg/μL, and 1.0 pg/μL). Both RT-PCR amplifications were performed under the optimized multiplex RT-PCR assay conditions using the corresponding primers of the target fragments to be assayed.
Multiplex RT-PCR assays of field samples
During 2018–2020, a total of 59 mature passion fruit leaf samples with symptoms such as yellowing, mosaic, or with no visible symptoms were collected from orchards of Xunwu, Ruijin, and Yudu counties in Jiangxi Province, China. The sampled passion fruit plants were all cultivar of purple passion fruit and planted in the same year. Total RNA was extracted from the 59 field samples as described in “Plants and viruses”. The RNA from an identified virus-negative field passion fruit sample was used as a negative control, and RNA from the virus source passion fruit plant was used as a positive control. The multiplex assay results of the field samples were verified using simplex RT-PCR assays. All the above experiments were carried out three times independently to ensure reproducibility and eliminate experimental differences.
Results
Specificity and compatibility of primers
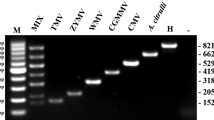
In simplex RT-PCR, the expected sizes of viral target genes for CiaRV (597 bp), EAPV (529 bp), PLV (320 bp), and TeMV (235 bp) were amplified specifically for all four primer pairs (Fig. 2, Lane 2–5). For multiplex RT-PCR, the primer combinations also gave clear and specific bands corresponding to target products. The amplified PCR products were sequenced and submitted to GenBank (GenBank accession No. OP328809, OP328810, OP328811, OP328812). These sequences were also subjected to BLASTn search to determine their homology and showed the highest sequence identity to CiaRV, EAPV, PLV, and TeMV isolates (GenBank accession No. MT302542, MT450870, MH379331, MK340754), respectively. The results further confirmed that the correct fragments had been amplified. The primer combinations were used for further optimization in multiplex RT-PCR (Fig. 2, Lane 1). No significant primer interaction or primer-dimer formation was observed (Fig. 2, Lane N).
Agarose gel analysis of the specificity of four primer pairs used for the simplex and multiplex RT-PCR assay for CiaRV, EAPV, PLV, and TeMV. Lane 1: multiplex RT-PCR of all four viruses; Lane 2: simplex RT-PCR for CiaRV (597 bp); Lane 3: simplex RT-PCR for EAPV (529 bp); Lane 4: simplex RT-PCR for PLV (320 bp); Lane 5: simplex RT-PCR for TeMV (235 bp); Lane N: Negative control. Lane M: TaKaRa DL 2000 DNA marker
Optimization and establishment of multiplex RT-PCR
Different amplification efficiencies indicated by different band intensities were obtained when equal concentrations (0.15 pM) of the four primer pairs were used in multiplex RT-PCR (Fig. 3A, Lane 1). To optimize the multiplex PCR system, combinations of different concentrations of the four primer pairs were tested in multiplex RT-PCR. The results showed that the amplification efficiencies for EAPV varied with different combinations of primer concentrations and that those for CiaRV, PLV, and TeMV remained stable. Balanced amplification was achieved when the primer concentrations were 0.3 pM for EAPV and 0.15 pM for CiaRV, PLV, and TeMV (Fig. 3A, Lane 4).
Agarose gel analysis of the products of the multiplex RT-PCR optimization experiments. A Optimization of the concentrations of four primer pairs. Lanes 1–4 indicate the concentration combinations of four primer pairs specific for CiaRV, EAPV, PLV, and TeMV: (1) 0.15:0.15:0.15:0.15 pM; (2) 0.15:0.20:0.15:0.15 pM; (3) 0.15:0.25:0.15:0.15 pM; (4) 0.15:0.30:0.15:0.15 pM. B Optimization of RT-PCR annealing temperature. Lanes 1–8: annealing at 52.0 ℃, 53.0 ℃, 54.0 ℃, 55.0 ℃, 56.0 ℃, 57.0 ℃, 58.0 ℃ and 59.0 ℃, respectively. C Optimization of RT-PCR extension time and amplification cycles. Lanes 1–3: extension times of 45 s, 60 s, and 75 s, amplification cycles of 30; Lanes 4–6: extension times of 45 s, 60 s, and 75 s, amplification cycles of 35; Lanes 7–9: extension times of 45 s, 60 s, and 75 s, amplification cycles of 40. Lane M: TaKaRa DL 2000 DNA marker
Gradient RT-PCR was performed to determine the optimal annealing temperature. When the annealing temperature was set as 56 °C, it obtained the best amplification result. Therefore, 56 °C was used as the optimal annealing temperature for the multiplex RT-PCR (Fig. 3B). In testing various combinations of extension time and number of cycles, the results were obtained when the extension time was 60 s and the cycle number was 35 (Fig. 3C).
Sensitivities of the simplex and multiplex RT-PCR assays
The results showed that the detection limits of the respective simplex RT-PCR for CiaRV were equivalent to 100 pg of total RNA, those for EAPV and PLV were 10 pg of total RNA, and those for TeMV were 1.0 ng of total RNA (Fig. 4A–D). On the other hand, in the multiplex RT-PCR assay, the detection limit was 100 pg of total RNA for CiaRV and EAPV, 10 pg of total RNA for PLV, and 1.0 ng of total RNA for TeMV (Fig. 4E). These results indicated that multiplex RT-PCR was only tenfold less sensitive than simplex RT-PCR for the detection of EAPV.
Comparison of the sensitivities of the simplex RT-PCR (A–D) and multiplex RT-PCR (E) assays for the detection CiaRV, EAPV, PLV, and TeMV using tenfold serial dilutions of the total RNA template prepared from the virus source plant co-infected with all the four viruses. Lane M: TaKaRa DL 2000 DNA marker; Lane 1–6: tenfold serial dilutions of the total RNA (100 ng/μL, 10 ng/μL, 1.0 ng/μL, 100 pg/μL, 10 pg/μL, 1.0 pg/μL)
Evaluation of multiplex RT-PCR using field samples
The multiplex RT-PCR was validated for practical application by detecting CiaRV, EAPV, PLV, and TeMV using 59 field samples (Table 2). CiaRV, EAPV, PLV, and TeMV were detected in 4/59 (6.8%), 0/59 (0.0%), 0/59 (0.0%), and 26/59 (44.1%) samples, respectively. Double-virus mixed infections of CiaRV + TeMV or EAPV + TeMV were detected in 10/59 (16.9%). Triple-virus mixed infections of CiaRV + EAPV + TeMV or EAPV + PLV + TeMV were detected in 18/59 (27.1%). Quadruple-virus mixed infections of CiaRV + EAPV + PLV + TeMV were 1/59 (1.7%). Among the 59 samples, only 2 samples were not infected with any of the four viruses. The samples from Xunwu County had a high virus detection rate. All of them were infected with at least one virus, and 21 (95.5%) samples had mixed infections with 2–4 viruses (Fig. 5). All of the field-collected samples were further confirmed using simplex RT-PCR, and the results were in agreement with those of the multiplex RT-PCR, indicating that the multiplex RT-PCR assay developed in this study is suitable for the purpose of field sample detection.
Discussion
Passion fruit is an economically important fruit crop, and the planting area of passion fruit in southern China has gradually increased in recent years. Viral diseases are a major constraint for passion fruit production (Baker et al. 2011), and viruses mixed infection often caused more severe symptoms. Thus, the development of reliable, fast, and cost-effective detection method for the viral disease management remains a necessity. This paper describes a multiplex RT-PCR assay for the simultaneous detection of four viruses infecting passion fruit (Baker et al. 2011). The assay described above provides a sensitive tool for the simultaneous detection of CiaRV, EAPV, PLV, and TeMV in single or mixed infections and was successfully tested on 59 field samples.
One frequent problem encountered in multiplex RT-PCR is the unbalanced amplification of viruses due to the presence of multiple gene targets in one reaction and to primers conferring compatibility with their targets, which may result in competition for enzymes and nucleotides (Wei et al. 2008). In our study, weak amplification of the EAPV fragment was obtained when using equal concentrations of the four primer sets in the multiplex RT-PCR assay. To balance the amplification efficiency and ensure similar band intensities of the four viruses, eventually, relatively higher primer concentrations (0.30 pM) for EAPV were used for the production of similar intensities of the amplicons in the same reaction.
In addition, the sensitivity of the multiplex RT-PCR compared to that of simplex RT-PCR was approximately tenfold lower for the detection of EAPV. Similar findings have been reported in other studies (Wei et al. 2009; Xue et al. 2021; Yao et al. 2014), which is due to some factors affecting multiplex RT-PCR efficiency, such as high oligonucleotide and primer concentrations.
To validate the reliability of the multiplex RT-PCR assay in field applications, 59 passion fruit field samples were collected from three counties in the Gannan district of China and tested by the multiplex RT-PCR assay. The results of the multiplex RT-PCR assay indicated that the single infection rates of the four viruses were 4/59 (6.8%), 0/59 (0.0%), 0/59 (0.0%), and 26/59 (44.1%), respectively. TeMV had the highest detection rate. Furthermore, 27/59 (45.8%) of the samples were coinfected with 2–4 viruses. In the 29 coinfected samples, TeMV was also detected in each sample. The TeMV total detection rate was 53/59 (89.8%). According to this result, TeMV has a high prevalence and is the major passion fruit virus pathogen of Gannan district. Additionally, 57/59 (96.6%) of the collected samples were infected with at least one virus. These results suggested that virus infection may be very frequent in this passion fruit production area and represent a significant threat to passion fruit production. Passion fruit growers likely face important challenges and must take appropriate precautions to manage the spread of the viral disease.
To our knowledge, this is the first report of the simultaneous detection of CiaRV, EAPV, PLV, and TeMV using a multiplex RT-PCR assay. The optimized multiplex RT-PCR assay developed in this study is a convenient and sensitive method that can be used to detect coinfections of CiaRV, EAPV, PLV, and TeMV in diseased fruit plants. This assay should be more time and cost saving, especially for detecting coinfecting viruses in large-scale surveys, than the previous single-virus detection methods for these four viruses.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Baker CA, Jeyaprakash A, Webster CG, Adkins S (2011) Viruses infecting passiflora species in Florida. European Biophysics Journal 40:175–180
Chen SS, Yu NN, Yang SH, Zhong BP, Lan HH (2018) Identification of Telosma mosaic virus infection in Passiflora edulis and its impact on phytochemical contents. Virology Journal 15:168
Cheng YH, Deng TC, Chen CC, Chiang CH, Chang CA (2014) First report of Euphorbia leaf curl virus and papaya leaf curl Guangdong virus on passion fruit in Taiwan. Plant Disease 98:1746
Chiemsombat P, Prammanee S, Pipattanawong N (2014) Occurrence of Telosma mosaic virus causing fruit severe mosaic disease in Thailand and immunostrip test for rapid virus detection. Crop Protection 63:41–47
Cho ID, Yang CY, Yoon JY, Kwon TR, Hammond J, Lim HS (2021) First report of Passiflora latent virus infecting persimmon (Diospyros kaki) in Korea. Plant Disease 105:1236
Ferreira SS, Barros DR, de Almeida MR, Zerbini FM (2010) Characterization of passion fruit severe leaf distortion virus, a novel begomovirus infecting passion fruit in Brazil, reveals a close relationship with tomato-infecting begomoviruses. Plant Pathology 59:221–230
Fu X, Jiang J, Luo L, Du Q, Li X, Afandi A, Feng W, Xie X (2021) Development of reverse transcription loop-mediated isothermal amplification assay for rapid and visual detection of Telosma mosaic virus (TeMV) in passion fruit. Crop Protection 150:105795
Hao L, Xie JP, Chen SY, Wang SJ, Gong ZQ, Ling KS, Guo LY, Fan ZF, Zhou T (2016) A multiple RT-PCR assay for simultaneous detection and differentiation of latent viruses and apscarviroids in apple trees. Journal of Virological Methods 234:16–21
Huang AJ, Ding M, Cao MJ, Zhang S, Zhong LQ, Wang Y (2019) First report of papaya leaf curl China virus on passion fruit in China. Plant Disease 104:1265
Iwai H, Yamashita Y, Nishi N, Nakamura M (2006) The potyvirus associated with the dappled fruit of Passiflora edulis in Kagoshima prefecture, Japan is the third strain of the proposed new species East Asian Passiflora virus (EAPV) phylogenetically distinguished from strains of passion fruit woodiness virus. Archives of Virology 151:811–818
Jover-Gil S, Beeri A, Fresnillo P, Samach A, Candela H (2018) Complete genome sequence of a novel virus, classifiable within the Potyviridae family, which infects passion fruit (Passiflora edulis). Archives of Virology 163:3191–3194
Kawakami S, Morinaga M, Tsukamoto-Sen S, Mori S, Matsui Y, Kawama T (2021) Constituent characteristics and functional properties of passion fruit seed extract. Life (basel) 12:38
Noorani MS, Awasthi P, Sharma MP, Ram R, Zaidi AA, Hallan V (2013) Simultaneous detection and identification of four cherry viruses by two step multiplex RT-PCR with an internal control of plant nad5 mRNA. Journal of Virological Methods 193:103–107
Osman F, Hodzic E, Kwon SJ, Wang JB, Vidalakis G (2015) Development and validation of a multiplex reverse transcription quantitative PCR (RT-qPCR) assay for the rapid detection of Citrus tristeza virus, Citrus psorosis virus, and Citrus leaf blotch virus. Journal of Virological Methods 220:64–75
Pares RD, Gunn LV, Keskula EN, Martin AB, Teakle DS (2007) Occurrence of Passiflora latent carlavirus in cultivated and wild Passiflora species in Australia. Plant Disease 81:348–350
Sokhandan N, Gillings MR, Bowyer JW (1997) Polymerase chain reaction detection and assessment of genetic variation in New South Wales isolates of passion fruit woodiness potyvirus. Australasian Plant Pathology 26:155–164
Spiegel S, Zeidan M, Sobolev I, Beckelman Y, Holdengreber V, Tam Y, Joseph MB, Lipsker Z, Gera A (2007) The complete nucleotide sequence of Passiflora latent virus and its phylogenetic relationship to other carlaviruses. Archives of Virology 152:181–189
Tang J, Clover GRG, Alexander BJR, Quinn BD (2008) First report of Passiflora latent virus in banana passionfruit (Passiflora tarminiana) in New Zealand. Plant Disease 92:486
Thokchom R, Mandal G (2017) Production preference and importance of passion fruit (Passiflora Edulis): a review. Journal of Agricultural Engineering and Food Technology 4:27–30
Tuo DC, Shen WT, Yang Y, Yan P, Li XY, Zhou P (2014) Development and validation of a multiplex reverse transcription PCR assay for simultaneous detection of three papaya viruses. Virus 6:3893–3906
Wei T, Lu G, Clover GRG (2008) Novel approaches to mitigate primer interaction and eliminate inhibitors in multiplex PCR, demonstrated using an assay for detection of three strawberry viruses. Journal of Virological Methods 151:132–139
Wei T, Lu G, Clover GRG (2009) A multiplex RT-PCR for the detection of potato yellow vein virus, tobacco rattle virus and tomato infectious chlorosis virus in potato with a plant internal amplification control. Plant Pathology 58:203–209
Xie LX, Zhang XY, Zheng S, Zhang LJ, Li T (2017) Molecular identification and specific detection of Telosma mosaic virus infecting passion fruit. Scientia Agriculture Sinica 50:4725–4734
Xie LX, Zhang LJ, Zhang XY, Zheng S, Li T (2018) First report of East Asian passiflora virus infecting Passiflora edulis in Fujian. China Acta Horticulturae Sinica 45:1578–1594
Xue B, Shang J, Yang J, Zhang L, Du JB, Yu L, Yang WW, Naeem M (2021) Development of a multiplex RT-PCR assay for the detection of soybean mosaic virus, bean common mosaic virus and cucumber mosaic virus in field samples of soybean. Journal of Virological Methods 298:114278
Yao B, Wang G, Ma X, Liu W, Tang H, Zhu H, Hong N (2014) Simultaneous detection and differentiation of three viruses in pear plants by a multiplex RT-PCR. Journal of Virological Methods 196:113–119
Yu Y, Zhao Z, Jiang D, Wu Z, Li S (2013) A one-step multiplex RT-PCR assay for simultaneous detection of four viruses that infect peach. Letters in Applied Microbiology 57:350–355
Yu CW, Lian Q, Lin HH, Chen L, Lu YZ, Zhai YY, Han X, Du ZG, Gao FL, Wu ZJ (2021) A clade of telosma mosaic virus from Thailand is undergoing geographical expansion and genetic differentiation in passionfruit of Vietnam and China. Phytopathology Research 3:24
Zhang S, Huang AJ, Zhou X, Li ZH, Dietzgen RG, Zhou CY, Cao MJ (2020) Natural defect of a plant rhabdovirus glycoprotein gene: a case study of virus–plant coevolution. Phytopathology 111:227–236
Author information
Authors and Affiliations
Contributions
The author Aijun Huang devised the experiment and drafted the manuscript. Peipei Gu, Ying Wang, Jiali Li, and Zhixun Yang performed the experiments. The corresponding author Long Yi did critical revision of drafted manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, A., Gu, P., Wang, Y. et al. Simultaneous detection and differentiation of four viruses in passion fruit plants by a multiplex RT-PCR. Trop. plant pathol. 48, 23–29 (2023). https://doi.org/10.1007/s40858-022-00538-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-022-00538-5