Abstract
Purpose of Review
Organoids are an emerging technology utilizing three-dimensional (3D), multi-cellular in vitro models to represent the function and physiological responses of tissues and organs. By using physiologically relevant models, more accurate tissue responses to viral infection can be observed, and effective treatments and preventive strategies can be identified. Animals and two-dimensional (2D) cell culture models occasionally result in inaccurate disease modeling outcomes. Organoids have been developed to better represent human organ and tissue systems, and accurately model tissue function and disease responses. By using organoids to study SARS-Cov-2 infection, researchers have now evaluated the viral effects on different organs and evaluate efficacy of potential treatments. The purpose of this review is to highlight organoid technologies and their ability to model SARS-Cov-2 infection and tissue responses.
Recent Findings
Lung, cardiac, kidney, and small intestine organoids have been examined as potential models of SARS-CoV-2 infection. Lung organoid research has highlighted that SARS-CoV-2 shows preferential infection of club cells and have shown value for the rapid screening and evaluations of multiple anti-viral drugs. Kidney organoid research suggests human recombinant soluble ACE2 as a preventative measure during early-stage infection. Using small intestine organoids, fecal to oral transmission has been evaluated as a transmission route for the virus. Lastly in cardiac organoids drug evaluation studies have found that drugs such as bromodomain, external family inhibitors, BETi, and apabetalone may be effective treatments for SARs-CoV-2 cardiac injury.
Summary
Organoids are an effective tool to study the effects of viral infections and for drug screening and evaluation studies. By using organoids, more accurate disease modeling can be performed, and physiological effects of infection and treatment can be better understood.
Similar content being viewed by others
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged late 2019 and quickly became a worldwide pandemic. Spread through transmission of respiratory aerosols and droplets, its high infectious rate led to rapid illness, multi-organ disease, and deaths or long-term post-COVID sequelae. The high transmissibility and virulence of this virus highlighted the urgent need for physiologically relevant research models to study the interaction of the SARS-CoV-2 virus across multiple tissues, and identify novel therapeutics and treatment modalities.
There are currently an array of in vivo and in vitro model systems used to study the mechanisms of viral infections and evaluate novel drug candidates and therapies. These models range from simple monotypic cell cultures of immortalized cell lines to complex models that utilize entire organs or large animal species (Fig. 1). Any model system has their own strengths and limitations. For example, monotypic cultures of immortalized cell lines provide a cost-effective means for high-throughput assessment of virus infectivity, replication, or drug effects. With the average cost of developing a new drug ranging from $314 million to $2.8 billion, immortalized monotypic cultures provide a costly benefit [1]. However, immortalized cell lines are inherently abnormal in their genetic composition and generally lack physiologically accurate form, function, and complexity of their host tissues. These molecular differences may lead to research findings that are not reproducible in normal non-immortalized cells and tissues. Model limitations add to the median cost per product ($985 million between 2009 and 2018) for the research and development of new therapeutic drugs and biologic agents, including failed trails [1]. Clinical drug development trials have been estimated to have a 90% failure rate [2]. Total capitalized costs have been shown to increase at 7.4% annually, accounting for price inflation[3]. Improved model development can discover potential obstacles prior to the capital investment.
Of the available model systems, 2D cell culture models (tissue-specific monotypic cultures) are typically used to study the human response to viral infection on a cellular level. 2D models generally utilize a single well-defined cell type harvested from tissue cell isolation techniques, or cell lines which can be generated by a variety of means to enable expansion of these cell types. The isolated cells are typically expanded and passaged on tissue-coated plastic, collagen-coated plates, or Transwell membranes. Tissue-coasted plastic is the simplest form, relying on the cells ability to attach to plastic-based materials under submerged culture media incubation. Collagen coating of plastic plates is often performed to increase the cell attachment of less adherent cells, thus improving expansion efficiency. Transwell systems add complexity by allowing an air liquid interface for the monolayer, permitting barrier interactions and ion transport through a membrane into media. Transwell cultures are particularly useful for analyzing lung and airway cell cultures, since physiologically these cells are in an air-exposed environment. 2D models are useful for testing wide ranges of genetic variability based on the cell donors, with low maintenance needs and moderate to low cost [4]. Genetically engineered cell lines such as wild-type VeroE6 have been used for the successful isolation and propagation of the SARS-CoV-2 virus, as well as investigating the roles of certain enzymes in response to SARS-CoV-2 [5]. Studies have also shown viral replication in Vero- VeroE6 cells under trypsin-free environments [6]. Unfortunately, 2D models have many limitations, primarily due to their monotypic cell environment and lack of supporting tissue components that are present in vivo such as extracellular matrix components. 2D monotypic culture models are often more susceptible to treatments compared to humans or animal models, and their abilities to accurately model overall tissue function and responses are limited compared to more complex 3D co-culture models. 2D cultures often lack the complexity of tissue and diversity of cells in the individual model combined with limited lifespan in culture.
Unlike 2D model systems, animal models provide a complete functional organism that can be evaluated for systemic, tissue-specific responses to a virus, drug, or treatment. However, animal studies can be prohibitively expensive and, although animal models provide a systematic response, differences in animal anatomy, physiology, and immune responses often do not always reproduce the viral responses or pathogenesis observed in humans. In the field of oncology, for example, animal models’ average rate of useful translation to clinical trials is less than 8% [7]. Animal models tend to overestimate treatment effectiveness by about 30% due to negative results often not published[8]. Data suggests that between 41 and 89% of transcription factors among 4000 genes differ between humans and mice[9]. Along with physiological differences, animal model methodologies can also effect results. Stress is a common factor that may have significant effects on experimental results as well as therapeutic agents[10]. Preclinical trials have expanded on the correlation between animal model’s toxicity and observed human toxicity. Using data for over 108 drugs ranging from small molecules, biologics as well as conjugates showed a weak correlation for predictability with a median positive predictor value of 0.65 and a negative predictor value of 0.5 [11]. This correlation can be further examined when looking into animal models for SARS-CoV-2. For example, numerous studies have shown that SARS-CoV-2 targets angiotensin converting enzyme II (ACE2) for infection in humans, but the ACE2 receptors of wild-type mice prevent SARS-CoV-2 from entering the cells [12]. The inhibition of entry is most likely attributed to the histidine located at 353 position, whereas human ACE2 contains a lysine that promotes improved virus-receptor interaction [13]. Unlike mice, Syrian hamsters have been successfully infected with SARS-CoV-2 and exhibited rapid breathing, weight loss, high viral load in the lungs, and spleen and lymphoid atrophy within the first week [4, 14]. Similarly, ferret models showed increased body temperatures, combined with viral replication and shedding, which led to airborne transmission between animals in this study [4]. Exposed felines showed respiratory tract inflammation, along with viral replication in the nose and throat combined with airborne transmission. Canines, on the other hand, did not demonstrate viral replication post exposure and appeared to be resistant to the virus similar to pigs, chickens, and ducks [15]. These examples demonstrate the variability in animal study outcomes and highlights both the value and limitations of in vivo model systems. Recently, there have been advances in the generation of “humanized” animal models in which animals are engrafted with human cells and tissues [16]. Humanized animal models improve the ability to predict human responses to virus and therapeutics. However, the current models are limited to specific host species, limited humanized components, and have a higher cost than non-humanized animal models.
Basics of organoid systems
Organoids are broadly defined as complex, multicellular, 3D in-vitro culture structures, derived from primary tissues or a stem cell source, which retain characteristic features of the original tissue [4]. However, the functional definition of an organoid varies in the literature and among researchers in the field. Organoids have been described as primary cell explants grafted onto 3D extracellular matrices, cell cultures consisting of different cell types derived from organ specific progenitor cells or stem cells, and 3D cell clusters of primary tissue capable of self-organization, self-renewal, and function similar to that of the source tissue [17].
Adult stem cells (ASC) derived from primary tissue or reprogramed differentiated cell types such as pluripotent stem cells (PSCs), embryonic stem cells (ESCs), or induced PSCs (iPSCs) have been deployed to generate organoids [18,19,20]. Under optimal growth conditions, these cells demonstrate inherent capacity to proliferate, self-organize, and develop specific features of an organ structure and function [21]. Broadly, organoids maintain a heterogeneous cell population, cell–cell and cell–matrix interactions, and sustain native gene, protein, and metabolic profile with microarchitectural and stromal cues. Many organoid types are optimized specially for disease modeling, drug discovery, drug testing, and personalized medicine [4, 22,23,24,25,26,27,28,29,30] (Fig. 2).
Higher complexity primary cell-derived organoids are being developed in the form of 3D tissue systems called “organ-tissue-equivalents” (OTEs). OTEs are commonly fabricated using tissue engineering principles, incorporating multiple cell types, biomimetic extracellular matrix substrates, and an organized microarchitecture that mimics the tissue source. OTEs can also be incorporated into microfluidic chips, allowing for systematic infections and analysis. These OTEs can encompass multiple cell types and when integrated with microfluidic chips they are typically referred to as an “organ-on-a-chip.” This characteristic is especially useful for studying the effects of drugs on unintended organs using a system of integrated OTEs known as a “body-on-a-chip.” Inclusion of immune organoid cultures allows for an immune response to a stimulant and better system modeling [31].
Lung organoid models
The lung is a highly complex branched organ with more than 40 distinct cell types tailoring its unique cellular architecture. The cell types include epithelial, vascular, stromal, and immune cells. The epithelial cells undergo distinct branching morphogenesis to form airway conducting bronchi, which opens into air sacs and alveolar space to facilitate gas exchange. Airway conducting bronchi is composed of ciliated cells and secretory cells (goblet and club cells), whereas alveolar epithelial lining comprises two distinct cell types: alveolar type II cells (AEC2s) and alveolar type I cells (AEC1s) [32]. Considering cellular heterogeneity within the respiratory system and the specialized functions of individual cell types, it is unlikely that conventional cell line-based in vitro models could recapitulate in vivo lung physiology comparable to differentiated primary airway cell cultures. Therefore, complex 3D lung organoids provide an excellent platform for research considering their ability to bio-mimic airway epithelial function and the lung microenvironment [32,33,34,35].
Lung spheroids
Spheroids are typically less complex forms of an organoid, consisting of an aggregation of cells. Spheroids can be formed from a cell suspension that aggregates into a compact sphere, clonal proliferation under low adherence conditions, partial dissociation of tissue followed by compaction and remodeling, and lastly a combination of cutting tissue and expanding under low-adherence conditions [36]. Spheroids do not require any scaffold or matrix to form, but are merged through cellular interactions. Ions and nutrients diffuse through the cellular channels into the core cells, emphasizing the importance of cellular transport [37]. Due to significance of cellular transportation, spheroids are limited in size based on the diffusion of nutrients often creating necrotic cores. Spheroids contain surface exposed cells as well as buried cells with various levels of oxygenation [38]. The necrotic core produces spheroids with similar characteristics to in vivo tumors leading to a focus on oncology-based research. Spheroids have been used to study invasion, metastasis, antineoplastic drugs, radioresponsiveness, and hypoxic effects [36].
Lung organoids
Airway and alveolar lung organoids can also be developed from pluripotent stem cell (PSCs), adult stem cell (ASCs) [35, 39, 40], or progenitor cells dissociated and isolated from donor lung tissue. PSC-derived lung organoids are developed by differentiating progenitor stem cells into mature lung epithelium. PSC differentiation protocols are continuously evolving with the goal of optimizing PSC lung organoids with adult lung cell heterogeneity [41,42,43,44,45]. PSC-derived organoids have provided a unique advantage to investigate lung disease and infections at various developmental stages [46,47,48]. On other hand, ASC-derived lung organoids are developed using isolated adult stem cells from various lung sites. These organoids can mitigate the technical difficulties of PSC cell differentiation. Basal cells are building units for most upper airway organoids (tracheospheres and bronchospheres). These basal cells self-renew and differentiate into ciliated cells and secretory cells [32]. AEC2 serves as adult stem cell units for alveolar organoids and upon any injury, AEC2s regenerate and differentiate into AEC1s [49]. Conversely, ASC-derived lung organoids are used to investigate mature stages of various lung diseases especially lung cancer [50]. Due to ease of generation, ASC-derived lung organoids have enabled more rigorous tissue level research by providing high-throughput microengineering approaches.
Lung-on-a-chip
Primary cell-derived organoids in the form of lung-on-a-chip (LOC) models have been growing in interest due to their ability to accurately model air-tissue interactions. The lungs are a common point of entry for toxins, pathogens, and drugs through inhalation. LOC models can produce key aspects of physiological structure, including innate defense features like mucociliary transport. In addition to a differentiated airway epithelium, LOC models can contain a basement membrane and extracellular matrix, as well as supporting cell types, such as tissue fibroblasts, endothelial cells, and immune cells [51,52,53,54]. Recent advancements in LOC models incorporate pneumatic channels or diaphragm-like structures which induce cyclic wall stretching to simulate the mechanisms of breathing [55, 56]. These LOC models allow for aerosolized drug delivery to the epithelial surface and soluble drug delivery to the basolateral compartment to model intravenous drug delivery. The diversity of LOC models is increasing as they incorporate increasing complexity to achieve greater physiologic relevance. These systems hold promise for novel discoveries and increase the likelihood that research findings will be translatable and hold human relevance [31].
Other organoids relevant for SARS-CoV-2 disease modeling
Over recent years, there has been a rapid evolution and diversification of multiple organoid model systems including heart, brain, kidneys, and intestines. Like the respiratory system models, these organoids provide novel, physiologically relevant platforms for studying SARS-CoV-2 infection, COVID-19 sequelae, and emerging treatment strategies.
Brain organoids
SARS-CoV-2 has been linked to both short- and long-term neuropsychiatric symptoms and long-term brain sequelae in infected individuals [57]. Recent advancements in the development of complex brain organoids with multiple cell types may provide an improved model for elucidating the underlying mechanisms of COVID-19 pathologies. Embryonic stem cells or pluripotent stem cells, astrocytes, and neural progenitors all together tailor PSC-derived brain organoids. PSC-derived brain organoids are engineered to mimic specific regions (mini brain [58], ventral and dorsal forebrain [59, 60], choroid plexus [61], thalamus [62, 63], hypothalamus [64], midbrain [65, 66], and cerebellum [67]) of brain by guided differentiation [68]. Brain organoids have been used for models of neurological diseases, neuropsychiatric disorders, brain tumors, and drug testing in vitro [69, 70]. ASC-derived brain organoids are not preferred considering the surgical complications during tissue biopsy and complex cellular heterogeneity [40]. Development of blood brain barrier (BBB) 3D spheroids may be the solution to overcome these complications. Human spheroid models were generated containing human brain microvascular endothelial cells (HBMEC), human pericytes (HBVP), human astrocytes (HA), human microglia (HM), human oligodendrocytes (HO), and human neurons (HN). Combining these cell types with an endothelial cell core showed tight junction expression, adherens junctions, and cell-specific markers, suggesting a functional blood brain barrier model for drug screening and disease modeling [71].
Small intestine organoids
The small intestine is a major organ with specialized epithelium supported by mesenchyme which regulates digestive function and nutrient intake. The small intestines express ACE2 and TMPRSS2 in large quantities, suggesting a target for human enteric viruses [72]. Small intestinal organoids are generated from both PSC and ASC origins [73]. Small intestine organoids include enterocytes, goblet cells, enteroendocrine cells, transit amplifying cells, and stem cells [74]. Intestinal organoids play a vital role in understanding the changes within the intestinal microbiota during cancer and infectious diseases [75,76,77]. For instance, Heo et al. demonstrated propagation and complete life cycle of parasites within ASC-derived intestinal organoid, microinjected with Cryptosporidium parvum oocysts [78].
Cardiac organoids
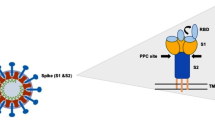
SARS-CoV-2 is known to affect many organs due to the presence of ACE2 and TMPRSS2 receptors throughout the body. In SARS-CoV-2 related deaths, there is a high association with myocardial injury [79]. Cardiac organoids generated from human pluripotent stem cells were shown to create chamber spaces as early as day 15, with gap junctions and tubular structures like the features found in early human fetal heart. The organoids consisted of 59% cardiomyocytes, 16% epicardial cells, 14% endocardial cells, 12% cardiac fibroblasts, and 1.6% endothelial cells, while producing beating for up to 8 weeks. These cardiac organoids were modified to resemble pregestational diabetes (PGD), exhibiting larger sizes reminiscent of macrosomia. The larger size suggests cardiac hypertrophy, reminiscence of PGD [80]. Other cardiac organoid models have been developed using induced pluripotent stem cells derived from cardiomyocytes, in the form of spheroids reaching about 250 µm in diameter. The spherical organoids were analyzed non-invasively for the change in beating kinetics as an indicator of health. The effects of epinephrine, isoproterenol, quinidine, astemizole, and ricin A chain were studied to verify the physiological relevance of the organoids. Isoproteerenol and epinephrine both increased the organoid beat rate to 40 beats per min and 42 beats per min, respectively. Whereas quinidine (12 beats per min), astemizole (4 beats per min), and ricing A chain (12 beats per min) reduced the beating rate, reminiscent of the observed physiological effects [81].
Kidney organoids
Viral components found in stool and urine also suggest SARS-CoV-2 may be able to affect the kidneys and small intestines. Kidney failure is a common disease observed from SARS-CoV-2 infections, but the mechanisms are still unknown [82]. Kidney organoids can be generated by creating a 2D culture containing wild-type iPSCs differentiated into mesoderm cells, then continuing differentiation into glomerular clusters and tubular structures. By day 18, the kidney organoids developed nephrogenic zones with discernible nephron-like structures. Using immunofluorescence, the localization of BM integrin receptors was confirmed, demonstrating kidney organoids mimic the normal development of kidney differentiation [83].
Application of organoids for biological and mechanistic viral studies
Organoids have been established as unique tools to better understand viral pathogenesis in ways that that are absent or non-physiological in 2D culture or animal models [84,85,86]. The application of organoids in viral research could have significant impacts on our ability to understand emerging viruses. In the twenty-first century alone, there has been a marked increase in viral epidemics including influenza A virus subtype H1N1 (H1N1) (2009), Middle East respiratory syndrome coronavirus (MERS-CoV) (2012), Ebola virus (2014), Zika virus (2015), and SARS-CoV-2 (2019) [87].
One of the most prominent early examples of the value of organoids for viral research was the use of brain organoids during the Zika virus outbreak. While the Zika virus is not a novel virus, the employment of human cerebral organoids provided the foremost opportunity to analyze fetal brain architecture and viral development in vitro [88,89,90,91]. Brain organoid research revealed many pathological characteristics of the Zika virus that influence its detrimental effects on developing fetal brains. Using forebrain-specific human iPSC organoids, Qian et al. demonstrated a microcephaly phenotype characteristic of Zika-infected fetal brains along with cellular tropism of neural progenitor cells [89]. More recently, Krenn et al. demonstrated virus-specific mechanisms with an attenuated interferon response characteristic of Zika microcephalic patients using Zika-infected brain organoids [92]. The organoid model highlighted cellular responses and distinct phenotypes not seen in 2D human neural progenitor cell cultures.
Other organoid models have been similarly used to study non-epidemic viruses. For instance, human intestinal organoids derived from primary tissue or stem cells have been crucial in understanding cell tropism for enteric viruses [93,94,95]. Prior to organoid models, it was near impossible to cultivate and replicate enteric viruses in the current intestinal cell lines. Similarly, Nie et al. developed iPSC-derived human liver organoids to illustrate and analyze hepatitis B viral infection and its associated viral life cycle that is not possible with other in vitro culture models and most animal models [96].
Lung organoid models have been used to investigate cellular tropism, viral replication, and pathogenesis of both seasonal respiratory viruses and cross-species epidemics viruses, such as avian H1N1 and H7N9 influenza [85, 97,98,99]. For instance, Bui et al. demonstrated successful infection and susceptibility of human lung organoids to seasonal influenza A and the less-studied influenza B viruses [98]. They were able to shed light on the pathogenicity of influenza B and the inhibitory effect of mucin on its viral replication. Another study using the same model examined the zoonotic potential of more recent strains of highly pathogenic avian H5N6/H5N8 influenza and showed lower replication competence indicative of low transmissibility [99]. Meanwhile, Harford et al. found that viruses like respiratory syncytial virus (RSV) can transfer hematogenously in utero to the developing fetal lungs [100]. Using a pluripotent stem cell lung organoid that models 1st trimester fetal development, they demonstrated that RSV causes significant changes to lung architecture and gene expression indicative of possible future chronic airway dysfunction.
Organoids in SARS-CoV-2 research
Lung organoids
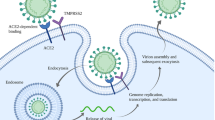
The respiratory epithelium is the primary site of SARS-CoV-2 infection and replication. Accordingly, there has been considerable interest in accurately modeling a functional respiratory epithelium with supporting tissues for COVID-19-related research (Table 1). Salahudeen and colleagues recently used single-cell transcriptomics on SARS-CoV-2-infected distal lung organoids to conclude that SCGB1A1+ club cells were a primary viral target [101]. The other studies report that SARS-CoV-2 targeted club cells on airway organoids with “apical-out” polarity and surface ACE2 expression [101]. Tissue-derived human bronchial epithelial organoids developed by Suzuki et al. showed enhanced expression of ACE2 and TMPRSS2 with differentiation in a Matrigel matrix. SARS-CoV-2 infection of these spheroid-like Matrigel organoids resulted in enhanced intracellular viral genome quantity, progeny virus, cytotoxicity, pyknotic cells, and increased type I interferon signals. Importantly, treatment of these organoids with the serine protease inhibitor Camostat significantly reduced viral copies [102]. Additional SARS-CoV-2 infection studies with lung organoids reported a significant reduction in type 1/3 interferon signaling post infection, which is consistent with infected patients [103]. Furthermore, studies comparing conducting airway and alveolar organoids identified sustained viral infection in the conducting airway cells, while distal alveolar organoids produced an immune response similar to that observed in fatal disease [104]. Pei et al. created human airway organoids and alveolar organoids using human embryonic stem cells with multiple differentiation stages. These differentiation stages started as embryonic stem cells, then proceeded to definitive endoderm, anterior foregut endoderm, ventralized anterior foregut endoderm, and lung progenitors before finalizing into human airway organoids and alveolar organoids. SARS-CoV-2 was shown to infect these organoids producing viral RNA as early as 24 h post infection. By using gene ontology analysis, it was confirmed that the genes downregulated were associated with cell and lipid metabolism, and the upregulated genes were associated with immune response. To protect against SARS-CoV-2, CB6, a neutralizing antibody proven to inhibit SARS-CoV-2 infection in rhesus monkeys, was analyzed. CB6 significantly repressed the generation of viral particles, in human organoids justifying the models used and drug candidate [105].
Brain organoids
There is suggestive evidence that SARS-CoV-2 may lead to short- and long-term neuropsychiatric symptoms indicative of independent brain damage. Although the brain is not the direct target of infection, SARS-CoV-2 may have the ability to pass through the blood brain barrier using circumventricular organs. Viral RNA has been detected in the medulla and cerebellum, as well as viral proteins found in brain vascular endothelium. Using brain organoids exposed to SARS-Cov-2, viral nucleoproteins were found in TUJ1-postive neurons and NESTIN neural progenitor cells. These organoids showed SARS-CoV-2 were able to enter the organoids, but not promote infection [106]. By utilizing human-induced pluripotent stem cell brain organoids, SARS-CoV-2 was proved to readily infect choroid plexus epithelial cells, while sparsely effecting neurons and astrocytes. Choroid plexus brain organoids showed increased inflammation, with altered barrier and secretory functions [107]. Mesci et al. created brain organoids using neural precursor cells, cortical neurons, and astrocytes. Using these brain organoids, sofosbuvir, an FDA approved anti-hepatitis C treatment, was tested as a possible treatment. Sofosbuvir was applied twice a week post infection, and successfully decreased the amount of neuronal cell death and viral accumulation as well as restoring protein expression in synaptic puncta. It is hypothesized that sofosbuvir can block replication by inhibiting SARS-CoV-2 RNA-dependent RNA polymerase. [108]
Small intestine organoids
In the intestines, ACE2 expression is substantially higher than other organs including the lungs [72]. Due to the high expression of ACE2 in the small intestines, possible fecal to oral transmission was speculated. For this to occur, a virus must survive the low pH, bile, and digestive enzymes combined with bacterial byproducts through the GI tract. SARS-CoV-2 lost effectiveness in simulated low pH gastric fluid; however, residual viruses were found in simulated human small intestine fluids containing biological surfactants [72]. Certain components in simulated human colonic fluids inactivated SARS-CoV-2 production [72]. Direct infections of human intestinal organoids derived from pluripotent stem cells showed SARS-CoV-2 sparring goblet cells lacking ACE2 expression. These organoids were used to study the effects of remdesivir on SAR-CoV-2 infections and concluded remdesivir inhibited infection at low micromolar concentrations. The model was also used to test the effect of famotidine, an over-the-counter histaimine-2 receptor antagonist, but showed relatively no effect on SARS-CoV-2 infections. EK1, a peptidic pancoronavirus fusion inhibitor, decreased viral infection by 38% 48 h after infection [109]. Small Intestine organoids have expanded the knowledge of infection and highlighted multiple ways to effect infections.
Cardiac organoids
The heart is also infected by SARS-CoV-2 due to the presence of ACE2 receptors. Patients with pre-existing cardiovascular diseases are more likely to die from an infection with 20–30% of hospitalized patients experiencing cardiac injury and dysfunction [79]. Using cardiac organoids with a co-culture of endothelial cells, a mixture of cytokines was identified as the inducing agent for diastolic dysfunction. Bromodomain and extraterminal family inhibitors decreased ACE2 expression and reduced infection of cardiomyocytes thus able to combat dysfunction. BETi and apabetalone may serve as effective treatments for SARS-CoV-2-mediated injury [110]. One of the ways to adapt organoids to focus on the mechanism of is by creating organoids with similar profiles to infected tissues. Human pluripotent stem cell–derived cardiac organoids were treated with IL-1 (beta), an upstream cytokine typically seen in SARS-CoV-2 infections, to induce cytokine storms that mirror the SARS-CoV-2 inflammatory profile. Using an inflammatory equivalent organoid, immunomodulatory drugs were analyzed for their effectiveness. IL-1RA (an IL-1 receptor antagonist), tocilizumab (an antibody against IL-6 receptor), baricitinib (a JAK/STAT inhibitor), and dexamethasone (a glucocorticoid) were applied to the cardiac organoids on day 0 and replenished at every media change. Dexamethasone was able to ameliorate the cardiac organoids, while tocilizumab and bricitinib were unable to improve contraction amplitude but preserved sarcomere width. Although dexamethasone was able to improve cardiac organoids, consequently it increased the presence of fibroblasts which may lead to further side effects [111].
Kidney organoids
Kidney organoids developed by Lin et al. displayed ACE2 receptors in the proximal tubule and podocyte II cell clusters, similar to native tissue [112]. The kidney organoids were generated from embryonic stem cells and differentiated producing tubular-like cells. Kidney organoids were infected and monitored for 6 days, afterwards expressing the ability to re-infect Vero E6 cells. The cells and tissues expressing the ACE2 receptors hosted viral replication, secreting infectious progeny virus within the supernatant. Kidney organoids led to the development of human recombinant soluble ACE2 that can block SARS-CoV-2 infections in the beginning phases [113]. This kidney model suggests that SARS-CoV-2 can infect the kidneys, but its contribution in multi-organ damage is unknown. Kidney organoids derived from human induced pluripotent stem cells expressed RNA sequences corresponding to the activation of profibrotic signaling pathways and overexpression of collagen 1 proteins when infected with SARS-CoV-2. In order to combat viral uptake in the kidney organoids, a noncovalent inhibitor of the SARS-CoV-2 main protease developed by the COVID Moonshot consortium was used and reduced intracellular SARS-CoV-2 RNA levels by over 4 logs [82]. This suggests protease inhibitors may be a potential treatment to reduce viral replication. The relationship between diabetes and SARS-CoV-2 was further expanded on using human pluripotent stem cell–derived kidney organoids. Diabetic kidney organoids were developed using high glucose oscillations and expressed increased collagen fiber deposition, and upregulated glycolysis-associated genes. By exposing infected organoids to an aerobic glycolysis inhibitors such as dichloroacetate (DCA), the organoids resulted in decreased infections providing insight on SARS-CoV-2 pathogenesis targeting metabolism [114].
Conclusion
Organoids have served as useful models to mimic the mechanisms and biological responses to SARS-CoV-2 infection and treatment. Organoid research has led to insight on the mechanisms of viral infection and disease and evaluation of possible treatment of SARS-CoV-2 infection. Through the development of more advanced and physiologically accurate organ and tissue-specific organoids, a better understanding of viral infection, disease development, and rapid screening and identification of efficacious treatments can occur. Organoids can take several forms ranging from spheroids to more complex microfluidic chip formats. Spheroids have been proven very useful for studying brain and cardiac effects, whereas 3D tissue organoids have been heavily used in the lungs and other organs. Organoids have identified the ACE2 receptor as the point of entry and identified a list of drug candidates including camostat and remdesivir, capable of prevention. Organoids provide more physiologically relevant models of human tissue function and disease, while enabling higher throughput and low costs. These features supplement current 2D and animal models and will enable acceleration of future advances in research across many fields.
References
Wouters OJ, McKee M, Luyten J. Estimated research and development investment needed to bring a new medicine to market, 2009–2018. JAMA. 2020;323(9):844–53. https://doi.org/10.1001/jama.2020.1166.
Sun D, Gao W, Hu H, Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharmaceutica Sinica B. 2022;12(7):3049–62. https://doi.org/10.1016/j.apsb.2022.02.002.
DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22(2):151–85. https://doi.org/10.1016/S0167-6296(02)00126-1.
Kim J, Koo B-K, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21(10):571–84. https://doi.org/10.1038/s41580-020-0259-3.
Chen D, Su X, Chen H, Chen S, Zhao Y, Wei W. Human organoids as a promising platform for fighting COVID-19. Int J Biol Sci. 2022;18(3):901–10. https://doi.org/10.7150/ijbs.64993.
Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease. United States Emerg Infect Dis. 2020;26(6):1266–73. https://doi.org/10.3201/eid2606.200516.
Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6(2):114–8.
Sena ES, van der Worp HB, Bath PMW, Howells DW, Macleod MR. Publication bias in reports of animal stroke studies leads to major overstatement of efficacy. PLoS Biol. 2010;8(3): e1000344. https://doi.org/10.1371/journal.pbio.1000344.
Johnson LSM. The trouble with animal models in brain research. In: Johnson LSM, Fenton A, Shriver A, editors. Neuroethics and Nonhuman Animals. Cham: Springer International Publishing; 2020. p. 271–86.
Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS. Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci Biobehav Rev. 2002;26(8):907–23. https://doi.org/10.1016/S0149-7634(02)00103-3.
Atkins JT, George GC, Hess K, Marcelo-Lewis KL, Yuan Y, Borthakur G, et al. Pre-clinical animal models are poor predictors of human toxicities in phase 1 oncology clinical trials. Br J Cancer. 2020;123(10):1496–501. https://doi.org/10.1038/s41416-020-01033-x.
••Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. https://doi.org/10.1038/s41586-020-2012-7. Findings from the study confimred the entry receptor for SARS-CoV-19 as the angiotensin converting enzyme (ACE2).
Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127-e220. https://doi.org/10.1128/JVI.00127-20.
Chan JF-W, Zhang AJ, Yuan S, Poon VK-M, Chan CC-S, Lee AC-Y, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–46. https://doi.org/10.1093/cid/ciaa325.
•Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–20. https://doi.org/10.1126/science.abb7015. Findings of this study highlight the physiological differeneces of SARs-CoV-19 infections in animal models.
Fan C, Wu Y, Rui X, Yang Y, Ling C, Liu S, et al. Animal models for COVID-19: advances, gaps and perspectives. Signal Transduct Target Ther. 2022;7(1):220. https://doi.org/10.1038/s41392-022-01087-8.
Simian M, Bissell MJ. Organoids: A historical perspective of thinking in three dimensions. J Cell Biol. 2017;216(1):31–40. https://doi.org/10.1083/jcb.201610056.
Shankaran A, Prasad K, Chaudhari S, Brand A, Satyamoorthy K. Advances in development and application of human organoids. 3 Biotech. 2021;11(6):257. https://doi.org/10.1007/s13205-021-02815-7.
Ashok A, Choudhury D, Fang Y, Hunziker W. Towards manufacturing of human organoids. Biotechnol Adv. 2020;39: 107460. https://doi.org/10.1016/j.biotechadv.2019.107460.
Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. 2021;6(5):402–20. https://doi.org/10.1038/s41578-021-00279-y.
Takebe T, Wells JM. Organoids by design. Science. 2019;364(6444):956–9. https://doi.org/10.1126/science.aaw7567.
Teriyapirom I, Batista-Rocha AS, Koo B-K. Genetic engineering in organoids. J Mol Med. 2021;99(4):555–68. https://doi.org/10.1007/s00109-020-02029-z.
Wagar LE, Salahudeen A, Constantz CM, Wendel BS, Lyons MM, Mallajosyula V, et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. 2021;27(1):125–35. https://doi.org/10.1038/s41591-020-01145-0.
Lehmann R, Lee CM, Shugart EC, Benedetti M, Charo RA, Gartner Z, et al. Human organoids: a new dimension in cell biology. Mol Biol Cell. 2019;30(10):1129–37. https://doi.org/10.1091/mbc.E19-03-0135.
Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246–54. https://doi.org/10.1038/ncb3312.
Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–18. https://doi.org/10.1038/s41568-018-0007-6.
Yoshida S, Miwa H, Kawachi T, Kume S, Takahashi K. Generation of intestinal organoids derived from human pluripotent stem cells for drug testing. Sci Rep. 2020;10(1):5989. https://doi.org/10.1038/s41598-020-63151-z.
Avnet S, Lemma S, Cortini M, Di Pompo G, Perut F, Baldini N. Pre-clinical models for studying the interaction between mesenchymal stromal cells and cancer cells and the induction of stemness. Front Oncol. 2019;9:305. https://doi.org/10.3389/fonc.2019.00305.
Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–97. https://doi.org/10.1016/j.cell.2016.05.082.
Li Y, Tang P, Cai S, Peng J, Hua G. Organoid based personalized medicine: from bench to bedside. Cell Regeneration. 2020;9(1):21. https://doi.org/10.1186/s13619-020-00059-z.
Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discovery Today. 2016;21(9):1399–411. https://doi.org/10.1016/j.drudis.2016.07.003.
Katsura H, Hogan BL. Lung organoids: powerful tools for studying lung stem cells and diseases. Lung Stem Cells in Development, Health and Disease (ERS Monograph) Sheffield, European Respiratory Society. 2021:175–89.
Nikolić MZ, Rawlins EL. Lung organoids and their use to study cell-cell interaction. Curr Pathobiol Rep. 2017;5(2):223–31. https://doi.org/10.1007/s40139-017-0137-7.
Kong J, Wen S, Cao W, Yue P, Xu X, Zhang Y, et al. Lung organoids, useful tools for investigating epithelial repair after lung injury. Stem Cell Res Ther. 2021;12(1):95. https://doi.org/10.1186/s13287-021-02172-5.
Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL. Lung organoids: current uses and future promise. Development. 2017;144(6):986–97. https://doi.org/10.1242/dev.140103.
Weiswald L-B, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17(1):1–15. https://doi.org/10.1016/j.neo.2014.12.004.
Konar D, Devarasetty M, Yildiz DV, Atala A, Murphy SV. Lung-on-a-chip technologies for disease modeling and drug development:supplementary issue: image and video acquisition and processing for clinical applications. Biomedical Engineering and Computational Biology. 2016;7s1:BECB.S34252. https://doi.org/10.4137/becb.S34252.
Page H, Flood P, Reynaud EG. Three-dimensional tissue cultures: current trends and beyond. Cell Tissue Res. 2013;352(1):123–31. https://doi.org/10.1007/s00441-012-1441-5.
Cunniff B, Druso JE, van der Velden JL. Lung organoids: advances in generation and 3D-visualization. Histochem Cell Biol. 2021;155(2):301–8. https://doi.org/10.1007/s00418-020-01955-w.
Shariati L, Esmaeili Y, Haghjooy Javanmard S, Bidram E, Amini A. Organoid technology: current standing and future perspectives. STEM CELLS. 2021;39(12):1625–49. https://doi.org/10.1002/stem.3379.
Dye BR, Hill DR, Ferguson MAH, Tsai Y-H, Nagy MS, Dyal R, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. https://doi.org/10.7554/eLife.05098.
Miller AJ, Dye BR, Ferrer-Torres D, Hill DR, Overeem AW, Shea LD, et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat Protoc. 2019;14(2):518–40. https://doi.org/10.1038/s41596-018-0104-8.
Gotoh S, Ito I, Nagasaki T, Yamamoto Y, Konishi S, Korogi Y, et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Reports. 2014;3(3):394–403. https://doi.org/10.1016/j.stemcr.2014.07.005.
Bhatia M. Current Protocols in stem cell biology. Current Protocols [Imprint]; 2015.
Kathiriya JJ, Wang C, Zhou M, Brumwell A, Cassandras M, Le Saux CJ, et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat Cell Biol. 2022;24(1):10–23. https://doi.org/10.1038/s41556-021-00809-4.
Tian L, Gao J, Garcia IM, Chen HJ, Castaldi A, Chen YW. Human pluripotent stem cell-derived lung organoids: Potential applications in development and disease modeling. Wiley Interdiscip Rev Dev Biol. 2021;10(6): e399. https://doi.org/10.1002/wdev.399.
van der Vaart J, Clevers H. Airway organoids as models of human disease. J Intern Med. 2021;289(5):604–13. https://doi.org/10.1111/joim.13075.
Chen Y-W, Huang SX, de Carvalho ALRT, Ho S-H, Islam MN, Volpi S, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol. 2017;19(5):542–9. https://doi.org/10.1038/ncb3510.
van Riet S, van Schadewijk A, Khedoe P, Limpens R, Bárcena M, Stolk J, et al. Organoid-based expansion of patient-derived primary alveolar type 2 cells for establishment of alveolus epithelial Lung-Chip cultures. Am J Physiol Lung Cell Mol Physiol. 2022;322(4):L526–38. https://doi.org/10.1152/ajplung.00153.2021.
Shi R, Radulovich N, Ng C, Liu N, Notsuda H, Cabanero M, et al. Organoid Cultures as preclinical models of non-small cell lung cancer. Clin Cancer Res. 2020;26(5):1162–74. https://doi.org/10.1158/1078-0432.Ccr-19-1376.
Konar D, Devarasetty M, Yildiz DV, Atala A, Murphy SV. Lung-on-a-chip technologies for disease modeling and drug development: supplementary issue: image and video acquisition and processing for clinical applications. Biomedical Engineering and Computational Biology. 2016;7s1:BECB.S34252. https://doi.org/10.4137/BECB.S34252.
Rajan SAP, Aleman J, Wan M, Pourhabibi Zarandi N, Nzou G, Murphy S, et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020;106:124–35. https://doi.org/10.1016/j.actbio.2020.02.015.
Skardal A, Aleman J, Forsythe S, Rajan S, Murphy S, Devarasetty M, et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. 2020;12(2): 025017. https://doi.org/10.1088/1758-5090/ab6d36.
Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep. 2017;7(1):8837. https://doi.org/10.1038/s41598-017-08879-x.
Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–8. https://doi.org/10.1126/science.1188302.
Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee H-H, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13(2):151–7. https://doi.org/10.1038/nmeth.3697.
Boldrini M, Canoll PD, Klein RS. How COVID-19 affects the brain. JAMA Psychiat. 2021;78(6):682–3. https://doi.org/10.1001/jamapsychiatry.2021.0500.
Tan HY, Cho H, Lee LP. Human mini-brain models. Nat Biomed Eng. 2021;5(1):11–25. https://doi.org/10.1038/s41551-020-00643-3.
Kim H, Xu R, Padmashri R, Dunaevsky A, Liu Y, Dreyfus CF, et al. Pluripotent stem cell-derived cerebral organoids reveal human oligodendrogenesis with dorsal and ventral origins. Stem Cell Reports. 2019;12(5):890–905. https://doi.org/10.1016/j.stemcr.2019.04.011.
Notaras M, Lodhi A, Barrio-Alonso E, Foord C, Rodrick T, Jones D, et al. Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol Psychiatry. 2021;26(12):7760–83. https://doi.org/10.1038/s41380-021-01189-9.
Xu H, Lehtinen MK. Choroid plexus organoids: harnessing csf gatekeepers for brain therapeutics. Cell Stem Cell. 2020;27(2):191–2. https://doi.org/10.1016/j.stem.2020.07.009.
Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24(3):487-97.e7. https://doi.org/10.1016/j.stem.2018.12.015.
Xiang Y, Cakir B, Park IH. Generation of regionally specified human brain organoids resembling thalamus development. STAR Protoc. 2020;1(1). https://doi.org/10.1016/j.xpro.2019.100001.
Huang WK, Wong SZH, Pather SR, Nguyen PTT, Zhang F, Zhang DY, et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 2021;28(9):1657-70.e10. https://doi.org/10.1016/j.stem.2021.04.006.
Smits LM, Schwamborn JC. Midbrain organoids: a new tool to investigate Parkinson’s disease. Front Cell Dev Biol. 2020;8:359. https://doi.org/10.3389/fcell.2020.00359.
Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports. 2017;8(5):1144–54. https://doi.org/10.1016/j.stemcr.2017.03.010.
Ballabio C, Anderle M, Gianesello M, Lago C, Miele E, Cardano M, et al. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat Commun. 2020;11(1):583. https://doi.org/10.1038/s41467-019-13989-3.
Jacob F, Schnoll JG, Song H, Ming GL. Building the brain from scratch: engineering region-specific brain organoids from human stem cells to study neural development and disease. Curr Top Dev Biol. 2021;142:477–530. https://doi.org/10.1016/bs.ctdb.2020.12.011.
Shou Y, Liang F, Xu S, Li X. The application of brain organoids: from neuronal development to neurological diseases. Front Cell Dev Biol. 2020;8: 579659. https://doi.org/10.3389/fcell.2020.579659.
Wang H. Modeling neurological diseases with human brain organoids. Front Synaptic Neurosci. 2018;10:15. https://doi.org/10.3389/fnsyn.2018.00015.
Nzou G, Wicks RT, Wicks EE, Seale SA, Sane CH, Chen A, et al. Human cortex spheroid with a functional blood brain barrier for high-throughput neurotoxicity screening and disease modeling. Sci Rep. 2018;8(1):7413. https://doi.org/10.1038/s41598-018-25603-5.
Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47). https://doi.org/10.1126/sciimmunol.abc3582.
Taelman J, Diaz M, Guiu J. Human intestinal organoids: promise and challenge. Front Cell Dev Biol. 2022;10: 854740. https://doi.org/10.3389/fcell.2022.854740.
Han Y, Yang L, Lacko LA, Chen S. Human organoid models to study SARS-CoV-2 infection. Nat Methods. 2022;19(4):418–28. https://doi.org/10.1038/s41592-022-01453-y.
Puschhof J, Pleguezuelos-Manzano C, Martinez-Silgado A, Akkerman N, Saftien A, Boot C, et al. Intestinal organoid cocultures with microbes. Nat Protoc. 2021;16(10):4633–49. https://doi.org/10.1038/s41596-021-00589-z.
Dutta D, Heo I, O'Connor R. Studying cryptosporidium Infection in 3D tissue-derived human organoid culture systems by microinjection. J Vis Exp. 2019(151). https://doi.org/10.3791/59610.
Min S, Kim S, Cho S-W. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp Mol Med. 2020;52(2):227–37. https://doi.org/10.1038/s12276-020-0386-0.
Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. 2018;3(7):814–23. https://doi.org/10.1038/s41564-018-0177-8.
Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, et al. Cardiovascular Implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol. 2020;5(7):811–8. https://doi.org/10.1001/jamacardio.2020.1017.
Lewis-Israeli YR, Wasserman AH, Gabalski MA, Volmert BD, Ming Y, Ball KA, et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat Commun. 2021;12(1):5142. https://doi.org/10.1038/s41467-021-25329-5.
Forsythe SD, Devarasetty M, Shupe T, Bishop C, Atala A, Soker S, et al. Environmental toxin screening using human-derived 3D bioengineered liver and cardiac organoids. Front Public Health. 2018;6:103.
Jansen J, Reimer KC, Nagai JS, Varghese FS, Overheul GJ, de Beer M, et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell. 2022;29(2):217-31.e8. https://doi.org/10.1016/j.stem.2021.12.010.
Morais MRPT, Tian P, Lawless C, Murtuza-Baker S, Hopkinson L, Woods S, et al. Kidney organoids recapitulate human basement membrane assembly in health and disease. eLife. 2022;11:e73486. https://doi.org/10.7554/eLife.73486.
Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353(6306):1387–93. https://doi.org/10.1126/science.aaf5211.
Zhou J, Li C, Sachs N, Chiu MC, Wong BH-Y, Chu H, et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proceedings of the National Academy of Sciences. 2018;115(26):6822–7. https://doi.org/10.1073/pnas.1806308115.
Driehuis E, Kolders S, Spelier S, Lõhmussaar K, Willems SM, Devriese LA, et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 2019;9(7):852–71. https://doi.org/10.1158/2159-8290.Cd-18-1522.
Morens DM, Fauci AS. Emerging pandemic diseases: how we got to COVID-19. Cell. 2020;182(5):1077–92. https://doi.org/10.1016/j.cell.2020.08.021.
Qian X, Nguyen HN, Jacob F, Song H, Ming GL. Using brain organoids to understand Zika virus-induced microcephaly. Development. 2017;144(6):952–7. https://doi.org/10.1242/dev.140707.
Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–54. https://doi.org/10.1016/j.cell.2016.04.032.
Pettke A, Tampere M, Pronk R, Wallner O, Falk A, Warpman Berglund U, et al. Broadly active antiviral compounds disturb zika virus progeny release rescuing virus-induced toxicity in brain organoids. Viruses. 2020;13(1). https://doi.org/10.3390/v13010037.
Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep. 2017;21(2):517–32. https://doi.org/10.1016/j.celrep.2017.09.047.
Krenn V, Bosone C, Burkard TR, Spanier J, Kalinke U, Calistri A, et al. Organoid modeling of Zika and herpes simplex virus 1 infections reveals virus-specific responses leading to microcephaly. Cell Stem Cell. 2021;28(8):1362-79.e7. https://doi.org/10.1016/j.stem.2021.03.004.
Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A. 2017;114(7):1672–7. https://doi.org/10.1073/pnas.1617363114.
Holly MK, Smith JG. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J Virol. 2018;92(9). https://doi.org/10.1128/jvi.00250-18.
Kolawole AO, Mirabelli C, Hill DR, Svoboda SA, Janowski AB, Passalacqua KD, et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019;15(10): e1008057. https://doi.org/10.1371/journal.ppat.1008057.
Nie YZ, Zheng YW, Miyakawa K, Murata S, Zhang RR, Sekine K, et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018;35:114–23. https://doi.org/10.1016/j.ebiom.2018.08.014.
Hui KPY, Cheung MC, Perera R, Ng KC, Bui CHT, Ho JCW, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8(7):687–95. https://doi.org/10.1016/s2213-2600(20)30193-4.
Bui CHT, Chan RWY, Ng MMT, Cheung MC, Ng KC, Chan MPK, et al. Tropism of influenza B viruses in human respiratory tract explants and airway organoids. Eur Respir J. 2019;54(2). https://doi.org/10.1183/13993003.00008-2019.
Bui CHT, Kuok DIT, Yeung HW, Ng KC, Chu DKW, Webby RJ, et al. Risk assessment for highly pathogenic avian influenza A(H5N6/H5N8) Clade 2.3.4.4 viruses. Emerg Infect Dis. 2021;27(10):2619–27. https://doi.org/10.3201/eid2710.210297.
Harford TJ, Rezaee F, Dye BR, Fan J, Spence JR, Piedimonte G. RSV-induced changes in a 3-dimensional organoid model of human fetal lungs. PLoS ONE. 2022;17(3): e0265094. https://doi.org/10.1371/journal.pone.0265094.
Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588(7839):670–5. https://doi.org/10.1038/s41586-020-3014-1.
•Suzuki T, Itoh Y, Sakai Y, Saito A, Okuzaki D, Motooka D, et al. Generation of human bronchial organoids for SARS-CoV-2 research. bioRxiv. 2020:2020.05.25.115600. https://doi.org/10.1101/2020.05.25.115600. Findings from this study evaluated camostat as a treatment option in human bronchial organoids.
Han Y, Yang L, Duan X, Duan F, Nilsson-Payant BE, Yaron TM, et al. Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv. 2020:2020.05.05.079095. https://doi.org/10.1101/2020.05.05.079095.
Tindle C, Fuller M, Fonseca A, Taheri S, Ibeawuchi SR, Beutler N, et al. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. Elife. 2021;10. https://doi.org/10.7554/eLife.66417.
Pei R, Feng J, Zhang Y, Sun H, Li L, Yang X, et al. Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell. 2021;12(9):717–33. https://doi.org/10.1007/s13238-020-00811-w.
Zhang B-Z, Chu H, Han S, Shuai H, Deng J, Hu Y-f, et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Research. 2020;30(10):928–31. https://doi.org/10.1038/s41422-020-0390-x.
Jacob F, Pather SR, Huang W-K, Zhang F, Wong SZH, Zhou H, et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27(6):937-50.e9. https://doi.org/10.1016/j.stem.2020.09.016.
Mesci P, Macia A, Saleh A, Martin-Sancho L, Yin X, Snethlage C, et al. Sofosbuvir protects human brain organoids against SARS-CoV-2. bioRxiv. 2020:2020.05.30.125856. https://doi.org/10.1101/2020.05.30.125856.
Krüger J, Groß R, Conzelmann C, Müller JA, Koepke L, Sparrer KMJ, et al. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol Gastroenterol Hepatol. 2021;11(4):935–48. https://doi.org/10.1016/j.jcmgh.2020.11.003.
Mills RJ, Humphrey SJ, Fortuna PRJ, Lor M, Foster SR, Quaife-Ryan GA, et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell. 2021;184(8):2167-82.e22. https://doi.org/10.1016/j.cell.2021.03.026.
Arhontoulis DC, Kerr C, Richards D, Tjen K, Hyams N, Jones JA, et al. Human cardiac organoids to model COVID-19 cytokine storm induced cardiac injuries. bioRxiv. 2022. https://doi.org/10.1101/2022.01.31.478497.
Lin W, Hu L, Zhang Y, Ooi JD, Meng T, Jin P, et al. Single-cell analysis of ACE2 expression in human kidneys and bladders reveals a potential route of 2019-nCoV infection. bioRxiv. 2020:2020.02.08.939892. https://doi.org/10.1101/2020.02.08.939892.
Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905-13.e7. https://doi.org/10.1016/j.cell.2020.04.004.
Garreta E, Prado P, Stanifer ML, Monteil V, Marco A, Ullate-Agote A, et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab. 2022;34(6):857-73.e9. https://doi.org/10.1016/j.cmet.2022.04.009.
Suezawa T, Kanagaki S, Korogi Y, Nakao K, Hirai T, Murakami K, et al. Modeling of lung phenotype of Hermansky-Pudlak syndrome type I using patient-specific iPSCs. Respir Res. 2021;22(1):284. https://doi.org/10.1186/s12931-021-01877-8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Sean Murphy has a patent US 11,001,811 B2 issued. The other authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Artificial Tissues
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sutton, K., Leach, T., Surendran, V. et al. Organoid Technologies for SARS-CoV-2 Research. Curr Stem Cell Rep 8, 151–163 (2022). https://doi.org/10.1007/s40778-022-00220-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-022-00220-1