Abstract
Wireless capsule endoscopy (WCE) might move through human body and captures the small bowel and captures the video and require the analysis of all frames of video due to which the diagnosis of gastrointestinal infections by the physician is a tedious task. This tiresome assignment has fuelled the researcher’s efforts to present an automated technique for gastrointestinal infections detection. The segmentation of stomach infections is a challenging task because the lesion region having low contrast and irregular shape and size. To handle this challenging task, in this research work a new deep semantic segmentation model is suggested for 3D-segmentation of the different types of stomach infections. In the segmentation model, deep labv3 is employed as a backbone of the ResNet-50 model. The model is trained with ground-masks and accurately performs pixel-wise classification in the testing phase. Similarity among the different types of stomach lesions accurate classification is a difficult task, which is addressed in this reported research by extracting deep features from global input images using a pre-trained ResNet-50 model. Furthermore, the latest advances in the estimation of uncertainty and model interpretability in the classification of different types of stomach infections is presented. The classification results estimate uncertainty related to the vital features in input and show how uncertainty and interpretability might be modeled in ResNet-50 for the classification of the different types of stomach infections. The proposed model achieved up to 90% prediction scores to authenticate the method performance.
Similar content being viewed by others
Introduction
An ulcer is a form of the gastrointestinal (GI) tract; about 10 percent of people have this condition. It is inflammatory chronic erosion or sore on the internal portion of the mucous skins [1, 2]. Itself ulcer is not fetal, but its symptoms are of serious ailments, i.e., Crohn’s ailment and the ulcerative of colitis might cause death at a complication stage [3]. Stomach ulcers are sores in the lining of the stomach and the duodenum. Up to 4 million peoples develop stomach ulcers in the United State per year (i.e., 1 out of 10 people) [4].
Conventional imaging protocols for ulcers are sonde and push endoscopy [5]. In the inspection process, it is entered into the anus or mouth of the patients by the experienced doctors to analyze the GI tract [6]. The traditional methodologies performed a vital role to analyze the lower and upper ends of GI [7]. Wireless capsule endoscopy (WCE) is an alternative method to offer painless, non-invasive, and direct small bowel inspection [8]. Commercially accessible WCE comprises the optical dome, part of illumination, batteries, and imaging sensors [9, 10]. WCE captures 2–4 images per second for nearly 8 h within the GI patient’s tract and transmits them wirelessly and placed in a machine connected to the patient’s waist [11]. Physicians can download and examine all photographs off-line for diagnostic purposes [12]. WCE creates approximately 55,000 images across each patient, in which 5% of images are normal from the whole collected WCE images, However, for physicians, it is a time-consuming and exhausting assignment [13]. Thus, it is important to develop an automated approach to analyze the ulcer images, and the physician’s workload is reduced [14]. The texture features [15] play an essential role in differentiation among the healthy/ulcer images. The Bidimensional EMD (BEEMD) method is used to classify the normal/ulcer images [16]. Curvelet-based lacunarity (DCT-LAC) technique, multi-level super-pixel approach are used for ulcer detection [17].
Detection of stomach infections at an initial stage may help to reduce the risk of mortality. Manual stomach infection evaluation is a laborious and time-consuming task in contrast to computerized methods used for the analysis of stomach. Stomach lesions segmentation and classification are performed using conventional and deep learning methodologies. Hand-crafted features are selected in classical approaches, whereas deep learning methods can learn to extract informative features in the pipeline. Although, a considerable amount of work has been done in this domain, while accurate stomach lesions detection is still a challenging [18]. This research work is based on two phases, in Phase-I deeplabv3 is utilized as a base model of the pre-trained ResNet-50 model to overcome the existing limitations. For accurate segmentation, the model is trained by selecting the hyperparameters after extensive experimentation. While, in Phase-II Resnet-50 model is trained using input images and classification outcome are analyzed using uncertainity based on the thresholding and Bayesian neural network to authenticate the prediction accuracy. The foremost contribution steps of the proposed model are as follows:
Phase I: Deeplabv3 with pre-trained ResNet-50 model for features mapping are developed for precise lesion segmentation. Phase II: The extracted features from ResNet-50 model and classification results are evaluated utilizing uncertainty by thresholding and BNN.
The organization article is manifested as follows: discussed related work in Sect. 2; proposed works is given in Sect. 3 and Sect. 4 explores experimental findings. In Sect. 5, conclusion is stated.
Related work
Much work has been devoted in developing an automated approach for ulcer detection [18,19,20,21,22]. Some latest existing techniques are discussed in this section. The stomach ulcer segmentation is a big challenge because endoscopy images having low contrast, illumination, and brightness issues, thus the infected region is not segmented accurately [23]. The classification of the different types of gastrointestinal infections is also an intricate task [24], because it relies on the feature’s extraction framework which directly impediment the classification accuracy [25]. An automated system has been presented to process the WCE images for early detection [26], where features are used and combined into a single vector and subsequently fed to the classifiers for ulcers/ bleeding classification [27]. This approach achieved an accuracy of 92.86% and 93.64% on bleeding and ulcers respectively [28]. Another approach has been presented for the detection of infection in the stomach and achieved an accuracy of 98.3% [29]. A method has been trained using the pre-trained ResNet-101 model and extracted features are optimized using the grasshopper method and passed to SVM for classification of different types of infections in the stomach such as a polyp, bleeding, and ulcer [25, 30]. The input images quality has been improved by applying a contrast enhancement and classical deep features [31] are extracted and best are selected using entropy [32,33,34]. These best features are passed as an input to the classifiers, in which KNN outperform as compared to other classifiers and achieved 99.42% accuracy [25]. The deep features have been extracted from transfer learning models such as AlexNet and Google Net for ulcer classification [35]. A saliency-based segmentation approach has been employed for ulcer segmentation [36]. The Hidden Markov Model (HMM) has been applied for the detection of stomach ulcers on two datasets [37]. An automated system has been presented which comprises the transformation of HSI, YIQ color [38, 39], and features fusion using singular value decomposition, [40] and finally classification is performed based on extracted features [41]. The square least saliency transformation with the probabilistic fitting model has been employed for the classification of stomach ulcers [42]. The weakly supervised neural model has been utilized for stomach ulcer detection. The extracted features from [43] VGG model and transferred as input to the classifiers for gastric ulcers classification [44]. Classical deep model has been utilized for stomach ulcer classification on 5560 images of WCE into ulcers, erosions/normal classes and its achieved accuracy of 90.8% [45]. The CNN model has been employed for the classification of different types of stomach lesions such as ulcers, bleeding and polyps with 72 and 71 percent specificity and sensitivity respectively [46].The GDP network has been utilized for stomach ulcer classification with 88.9% accuracy [47]. HA network with the residual model have been employed for stomach infection. The model achieved 91% accuracy [48].
In literature, extensive studies have been performed for the detection of different types of stomach infections; however, still, there is a gap in this domain because stomach lesions appear in a variable shape and size [14, 48, 58]. The selection of the learning parameters i.e., optimizing function, learning rate and batch-size of the CNN models is still a challenge that directly affects the classification accuracy. Pre-trained models such as Google net and Alex net are trained on the stomach ulcer datasets on 0.01 learning rate that does not provides satisfactory classification outcomes [35]. The MCNet does not provide accurate lesions segmentation due to unclear boundaries among the infected and the healthy regions [49].
Therefore, in this reported research a new framework is trained on optimum hyper-parameters for accurate segmentation. The deep extracted features from ResNet-50 model and supplied as an input to the softmax for stomach infections classification. Furthermore, uncertainty based on thresholding and Bayesian neural network [50] is performed to analyze the prediction scores.
Proposed methodology
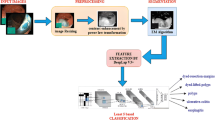
A modified model is presented for gastrointestinal infections detection. The technique comprises the two major phases as manifested in Fig. 1. In phase I, the infected stomach region is segmented with ground truth using a modified semantic segmentation model, whereas, a pre-trained ResNet-50 model is presented for the classification of different types of gastrointestinal infections such as Bleeding, Ulcer, Polyps, and normal stomach images.
Semantic segmentation of stomach ulcer
In the proposed model DeepLab v3 + network [51], is utilized as a bottleneck, in which CNN utilized encoder-decoder, skip connections, and dilated convolutions. The ResNet-50 is used as a head network of the deep labv3 for stomach infection segmentation. The semantic segmentation model comprises the 206 layers, which includes 01 input, 62 convolutional, 65 batch-normalization, 32 ReLU, 02 crop2d, 01 max-pooling, softmax, and pixel classification. The layered proposed semantic model is depicted in Fig. 2.
The hyperparameters for a proposed semantic segmentation model are given in Table 1.
Table 1, presents the model building hyperparameters such as 100 epochs, SGD optimizer, 0.001LR, and 16 batch- sizes are utilized for model training due to maximum accuracy. Figure 3, shows the segmented stomach lesions with ground masks.
Uncertainty estimation based on ResNet-50
In the medical domain, disease grading classification through computerized systems is much helpful for the gastroenterologist at the same time it has become complicated due to the increase in the size of the patient’s data. Currently, a convolutional neural network performs a vital role on larger datasets as compared to small-scale datasets. In this work ResNet-50 [52] is applied for model training which consists of 177 layers including 52 convolutional and batch-normalization, ReLU 49, 02 average pooling, 16 addition, 01 softmax, 01 classifications. Transfer learning is implemented for features mapping on stomach infection, which is previously trained on the ImageNet database. The features mapping is performed by the activation function of categorical cross-entropy which is defined as
where \({\mathrm{t}}_{\mathrm{i}}\) and \({\mathrm{s}}_{\mathrm{i}}\) denotes label and CNN score of each class(C). The 60:40 ratio is utilized for model training and testing. The description of the model with the number of layers and selected neurons are mentioned in Table 2.
The model training is performed on selected hyperparameters as mentioned in Table 3
Performance improvement via uncertainty-aware stomach infections classification
The uncertainty of the classification model is used for estimating the prediction in two ways (i) estimate the probability based on thresholding (ii) probability estimation based on the Bayesian neural network.
In this method, randomly complete data is split into training and testing parts. The threshold value is computed across each class label.
In BNN, given a dataset (D) =\({\mathrm{x}}_{\mathrm{n}}\in {\mathcal{R}}^{\mathrm{D}},{\mathrm{y}}_{\mathrm{n}}\in {\mathcal{R}}^{\mathrm{C}}{\}}_{\mathrm{n}=1}^{\mathrm{N}}\) where \({x}_{\mathrm{n}}\) represent input feature vector and \({\mathrm{y}}_{\mathrm{n}}\) denotes the one-hot encoded label vector. The predictive BNN on a new sampled \(\{x*,y*\}\) might be
where W represent weights, \(p\left(y*\left|x*,W\right.\right)\) denotes softmax function by \({f}_{\mathrm{W}}(x*)\) and \(p\left(W\left|x*,\right. D\right)\) shows posterior over weights. \(p\left(y*\left|x*,W\right.\right)\) shows network forward pass. The predictive distribution by Monte Carlo as defined as:
where predictive distribution might be computed through forward pass of a model T running with drop out employed to produce predictions T and computes standard deviation over softmax T samples outputs. The BNN utilized dropout for sampling to posterior predictive distribution that is referred as Monte Carlo dropout.
The predictions of all stomach infection test images are performed and sorted through their related uncertainty predictions. On the different uncertainty levels, predictions are conducted for diagnosis and compute the prediction accuracy at the specified threshold according to the class labels.
Dataset descriptions
In Table 4, proposed method performance is computed on five benchmark datasets such as a privately collected imaging dataset having 30 WCE videos, where 10 ulcer videos, 10 bleeding videos, and the remaining 10 videos are healthy [41]. Each video contains 500 frames. The CVC–Clinic DB database contains 612 WCE images with annotated ground truth [53]. The Nerthus dataset contains 21 WCE videos with 5525 number of frames [54]. The kvasir-segmentation dataset comprises 1000 WCE images with ground-masks [55]. The kvasir-classification dataset contains 4000 images of 8 classes, where each class contains 500 images of different types of stomach infections [56].
Experimental results and discussion
For evaluation of the efficiency of the proposed system, two experiments were carried out. Experiment#1 is done to compute the proposed segmentation model performance with ground annotated masks. Experiment#2 is implemented to analyze the classification results. The overall experiments are implemented on MATLAB 2020RA toolbox with coreI7 CPU, 32 GB RAM, and 8 GB Nvidia graphic card 2070 RTX.
Experiment#1 evaluation of semantic segmentation
The proposed semantic segmentation method performance is evaluated with ground annotated masks as given in Table 5.
The segmented stomach lesions achieved global accuracy of 0.98, 1.00, 0.98 on CVC–Clinic DB, Kvasir-SEG, and Private collected images respectively. The segmentation results with ground masks on benchmark datasets are shown in Figs. 4, 5, 6.
The segmented stomach lesions are computed with truth annotations masks on three benchmark datasets such as kvasir, private collected images, and CVC-CLINIC. Figures 4, 5, 6 shows that the proposed method more precisely segments the stomach infections. The proposed segmentation results are compared on the same benchmark datasets as mentioned in Table 6.
Table 6, shows the existing methodologies for segmentation of stomach infections such as [9, 41, 52, 53, 59, 66]. In the comparison analysis, the FCN method has been employed with 8, 16, and 32 fully connected layers, in which FCN-32 s achieved the highest 0.83, mean accuracy [57]. Seg-network [58] and dilation model [59] obtained 0.85 segmentation accuracy, while the U-net model [60] has been employed with different pre-trained networks such as VGG-16, VGG-19, and ResNet-34 [61]. Without any combination, only the U-net model achieved 0.86 mean segmentation accuracy, which is maximum compared to other pre-trained models. MCNet [62] model is employed for lesion segmentation with 0.84 mean accuracy. Comparison reflects that in the proposed model, deeplabv3 is used as a backbone of the ResNet-50 model and it has attained 0.98 mean accuracy which is also superior compared to recent all published work in this domain.
The proposed segmentation results are also compared with the U-net [60] model, the visually segmentation results as seen in Fig. 7.
Figure 7 results show that, on U-net segmentation model false positive rate is increased due to the segmentation of non-lesions pixels, while the proposed segmentation model (deeplabv3 & ResNet-50) segment the actual stomach lesions more precisely as compared to other models.
Experiment#2 classification of different types of stomach infections
The classification results on Nerthus-dataset-frames, private collected images, and kvasir classification dataset as given in Table 7, 8, 9, 10, 11, 12, 13.
Classification results on Nerthus-dataset-frames
The classification of four different types of stomach infections such as Grade-1 Bowl, Grade-2Bowl, Grade-3Bowl, and Grade-4Bowl. The classification results of different grading of the Bowl are analysed in terms of uncertainty measures such as thresholding and BNN. The proposed method achieved a 1.00 prediction rate on the uncertainty based on the thresholding. The prediction results are shown in Table 7 and Figs. 8, 9.
Table 7, shows the probability of the prediction scores based on thresholding, where overall achieved accuracy is 1.00. The precision rates of the Bowl grades are 1.00, 1.00, 0.99, 0.99 on Grade1, Grade 2, Grade3, and Grade 4 respectively. Similarly, in the same experiment, the prediction rate has computed using BNN as presented in Table 7 and Figs. 10, 11.
The prediction scores on BNN, the proposed method achieved maximum 0.96 accuracy and precision rate are 0.96, 0.97, 0.96, 0.96 on Grade Bowl-0, Grade Bowl-1, Grade Bowl-2, and Grade Bowl-3 respectively.
Classification results on kvasir-classification dataset
The classification results are computed on eight different kinds of stomach infections. The classification results are also computed on uncertainty-based thresholding and BNN as given in Table 9, 10 and Figs.12, 13.
The classification results on kvasir dataset using BNN as given in the Table 8 and Figs. 14, 15.
Classification results on private collected images
The classification results on private collected are manifested in Figs. 16, 17 and Table 11. The proposed method achieved testing accuracy of 0.982 and 0.10917 loss rates.
On private collected images, classification is performed into three classes such as bleeding, healthy, and ulcers. The method achieved cumulative accuracy of 1.00, however, the precision rate of each class obtained 1.00 on healthy, bleeding, and ulcer, respectively. Classification results on uncertainty based on BNN are illustrated in Figs. 18, 19 and Table 11.
On private collected images, BNN achieved 0.98 prediction scores on three classes such as healthy, bleeding, and ulcer. Similarly, 0.98 precision rate on healthy, bleeding, and 0.99 on ulcer class. The classification outcomes are compared with recent approaches as stated in Table 13.
The comparison of the proposed classification results is performed with eight existing methodologies, where features extraction, selection, and fusion approaches have been used for the classification of normal and bleeding WCE images with 0.94 accuracy. The pre-trained AlexNet model has been utilized for the classification of ulcer and erosion [63]. The classical Gabor features with pre-trained dense-Net has been employed for cancer/normal images classification [64]. Deep features are extracted from WCE images for classification of normal/ulcer [65]. The classical and deep features are fused and informative features are selected for Polyp, ulcer, esophagitis, bleeding, and normal images classification [22]. The duo-deep model has been employed for the classification of ulcer, polyp, bleeding [66]. StomachNetwork has been utilized for the classification of Polyps, Ulcer and normal images with 0.96 accuracy [67]. The proposed method classified the esophagitis, colon polyps, normal, bleeding, and UC (ulcers) images with the highest prediction scores compared to existing works.
After the comparison analysis, we conclude that, no methods exist in the literature for segmentation and classification of the different types of stomach infections by using all publically available challenging datasets and private collected images. This research work investigates a new approach for segmentation and classification of the different types of stomach infections, where kvasir-Seg, CVCClinicDB and private collected images with ground-masks are used to compute the segmentation model performance. Whereas for classification four classes (bowl-grade1, bowl-grade2, bowl-grade3, and bowl-grade4) of Nerthus dataset, eight classes of the kvasir dataset and three classes (polyp, bleeding and healthy) of the private collected images are utilized. The classification outcomes indicate that the proposed methodology is superior to current works as compared to existing current works, which authenticate the proposed method contribution.
Conclusion
In this research, a new approach is presented for analysis of stomach infections but It is a difficult job using WCE because lesions having irregular shapes and sizes. The informative features extraction is still a challenge because it’s reduced the classification accuracy. Accurate segmentation is performed using a deep semantic segmentation model, where deeplabv3 is employed as a bottleneck of the ResNet-50 model. The proposed modified model segments the stomach lesions in terms of different measures such as 0.97 meanIoU, 0.98 global accuracy, 0.96 weighted IoU, 0.98 mean accuracy and 0.98 mean BF-score on CVC–Clinic DB, whereas 1.00 meanIoU, 1.00 global accuracy, 1.00 weighted IoU, 1.00 mean accuracy and 1.00 mean BF-score on Kvasir-SEG, while 0.99 meanIoU, 0.98 global accuracy, 0.99 weighted IoU, 0.99 mean accuracy and 0.99 mean BF-score on Private collected images. The prediction scores of the classification models across each benchmark dataset are computed using uncertainty based on the standard thresholding and Bayesian neural network (BNN). The uncertainty based on thresholding the proposed approach attained an accuracy of 1.00 on private collected images and Nerthus dataset, while 0.96 using BNN on Nerthus frames. Similarly, on the same experiment, kvasir dataset achieved accuracy of 0.87 using uncertainty based on thresholding and 0.64 using uncertainty based on BNN. In the future accuracy on kvasir dataset might be further improved to enhanced the prediction rate of stomach infection.
References
Iijima K, Kanno T, Koike T, Shimosegawa T (2014) Helicobacter pylori-negative, non-steroidal anti-inflammatory drug: negative idiopathic ulcers in Asia. World J Gastroenterol WJG 20:706
Lapina PD What is the difference between erosion and stomach ulcers? What and how it hurts during erosion
Zonderman J (2000) Understanding Crohn disease and ulcerative colitis. Univ. Press of Mississippi, Mississippi
Harvard Health Publishing (2021) https://www.health.harvard.edu/digestive-health/peptic-ulcer-overview. Accessed by 07 Feb 2021
Appleyard M, Fireman Z, Glukhovsky A, Jacob H, Shreiver R, Kadirkamanathan S, Lavy A, Lewkowicz S, Scapa E, Shofti R (2000) A randomized trial comparing wireless capsule endoscopy with push enteroscopy for the detection of small-bowel lesions. Gastroenterology 119:1431–1438
Edwards L, Pfeiffer R, Quigley E, Hofman R, Balluff M (1991) Gastrointestinal symptoms in Parkinson’s disease. Mov Disord Off J Mov Disorder Soc 6:151–156
Abraham NS, Hartman C, Richardson P, Castillo D, Street RL Jr, Naik AD (2013) Risk of lower and upper gastrointestinal bleeding, transfusions, and hospitalizations with complex antithrombotic therapy in elderly patients. Circulation 128:1869–1877
Sohag MHA (2020) Detection of intestinal bleeding in wireless capsule endoscopy using machine learning techniques. University of Saskatchewan, Saskatchewan
Umay I, Fidan B, Barshan B (2017) Localization and tracking of implantable biomedical sensors. Sensors 17:583
Yuan Y, Li B, Meng MQ-H (2015) Bleeding frame and region detection in the wireless capsule endoscopy video. IEEE J Biomed Health Inf 20:624–630
Pan G, Wang L (2011) Swallowable wireless capsule endoscopy: progress and technical challenges. Gastroenterol Res Pract 2012
National Academies of Sciences E, Medicine (2015) Improving diagnosis in health care. National Academies Press, Washington
Suman S, Hussin FA, Nicolas W, Malik AS (2016) Ulcer detection and classification of wireless capsule endoscopy images using RGB masking. Adv Sci Lett 22:2764–2768
Hu H, Zheng W, Zhang X, Zhang X, Liu J, Hu W, Duan H, Si J (2020) Content-based gastric image retrieval using convolutional neural networks. Int J Imaging Syst Technol 31(1):439–449
Shabbir B, Sharif M, Nisar W, Yasmin M, Fernandes SL (2016) Automatic cotton wool spots extraction in retinal images using texture segmentation and gabor wavelet. J Integr Des Process Sci 20:65–76
Charisis V, Tsiligiri A, Hadjileontiadis LJ, Liatsos CN, Mavrogiannis CC, Sergiadis GD (2010) Ulcer detection in wireless capsule endoscopy images using bidimensional nonlinear analysis. In: XII mediterranean conference on medical and biological engineering and computing 2010. Springer, pp 236–239
Eid A, Charisis VS, Hadjileontiadis LJ, Sergiadis GD (2013) A curvelet-based lacunarity approach for ulcer detection from wireless capsule endoscopy images. In: Proceedings of the 26th IEEE international symposium on computer-based medical systems. IEEE, pp 273–278
Muhammad K, Khan S, Kumar N, Del Ser J, Mirjalili S (2020) Vision-based personalized wireless capsule endoscopy for smart healthcare: taxonomy, literature review, opportunities and challenges. Future Gener Comput Syst 113:266–280
Ali H, Sharif M, Yasmin M, Rehmani MH, Riaz F (2019) A survey of feature extraction and fusion of deep learning for detection of abnormalities in video endoscopy of gastrointestinal-tract. Artif Intell Rev 1:1–73
Liaqat A, Khan MA, Shah JH, Sharif M, Yasmin M, Fernandes SL (2018) Automated ulcer and bleeding classification from WCE images using multiple features fusion and selection. J Mech Med Biol 18:1850038
Liaqat A, Khan MA, Sharif M, Mittal M, Saba T, Manic KS, Al Attar FN (2020) Gastric tract infections detection and classification from wireless capsule endoscopy using computer vision techniques: a review. Curr Med Imaging 16:1229–1242
Majid A, Khan MA, Yasmin M, Rehman A, Yousafzai A, Tariq U (2020) Classification of stomach infections: a paradigm of convolutional neural network along with classical features fusion and selection. Microsc Res Tech 83:562–576
Kollias D, Tagaris A, Stafylopatis A, Kollias S, Tagaris G (2018) Deep neural architectures for prediction in healthcare. Complex Intell Syst 4:119–131
Gorbach S (1996) Chpter 95: Microbiology of the gastrointestinal tract. Medical microbiology, 4th edn. Baron S (eds.), University of Texas Medical Branch at Galveston, Galveston Retrieved from https://www.ncbi.nlm.nih.gov/
Sharif M, Attique Khan M, Rashid M, Yasmin M, Afza F, Tanik UJ (2019) Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images. J Exp Theor Artif Intell 1:1–23
Li B, Meng MQ-H (2012) Automatic polyp detection for wireless capsule endoscopy images. Expert Syst Appl 39:10952–10958
Rahim T, Usman MA, Shin SY (2020) A survey on contemporary computer-aided tumor, polyp, and ulcer detection methods in wireless capsule endoscopy imaging. Comput Med Imaging Graph 1:101767
Yeh J-Y, Wu T-H, Tsai W-J (2014) Bleeding and ulcer detection using wireless capsule endoscopy images. J Softw Eng Appl 7:422
Khan MA, Sharif M, Akram T, Yasmin M, Nayak RS (2019) Stomach deformities recognition using rank-based deep features selection. J Med Syst 43:329
Gamage H, Wijesinghe W, Perera I (2019) Instance-based segmentation for boundary detection of neuropathic ulcers through Mask-RCNN. In: International conference on artificial neural networks. Springer, pp 511–522
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Future Gener Comput Syst 87:290–297
Arunkumar N, Ramkumar K, Venkatraman V, Abdulhay E, Fernandes SL, Kadry S, Segal S (2017) Classification of focal and non focal EEG using entropies. Pattern Recogn Lett 94:112–117
Raja N, Rajinikanth V, Fernandes SL, Satapathy SC (2017) Segmentation of breast thermal images using Kapur’s entropy and hidden Markov random field. J Med Imaging Health Inf 7:1825–1829
Rajinikanth V, Satapathy SC, Fernandes SL, Nachiappan S (2017) Entropy based segmentation of tumor from brain MR images–a study with teaching learning based optimization. Pattern Recogn Lett 94:87–95
Alaskar H, Hussain A, Al-Aseem N, Liatsis P, Al-Jumeily D (2019) Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images. Sensors 19:1–16
Yuan Y, Wang J, Li B, Meng MQ-H (2015) Saliency based ulcer detection for wireless capsule endoscopy diagnosis. IEEE Trans Med Imaging 34:2046–2057
Khan MA, Sarfraz MS, Alhaisoni M, Albesher AA, Wang S, Ashraf I (2020) StomachNet: optimal deep learning features fusion for stomach abnormalities classification. IEEE Access
Nida N, Sharif M, Khan MUG, Yasmin M, Fernandes SL (2016) A framework for automatic colorization of medical imaging. IIOAB J 7:202–209
Yasmin M, Sharif M, Irum I, Mehmood W, Fernandes SL (2016) Combining multiple color and shape features for image retrieval. IIOAB J 7:97–110
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett
Khan MA, Rashid M, Sharif M, Javed K, Akram T (2019) Classification of gastrointestinal diseases of stomach from WCE using improved saliency-based method and discriminant features selection. Multimed Tools Appl 78:27743–27770
Kundu AK, Fattah SA, Wahid KA (2020) Least square saliency transformation of capsule endoscopy images for PDF model based multiple gastrointestinal disease classification. IEEE Access 8:58509–58521
Georgakopoulos SV, Iakovidis DK, Vasilakakis M, Plagianakos VP, Koulaouzidis A (2016) Weakly-supervised convolutional learning for detection of inflammatory gastrointestinal lesions. In: 2016 IEEE international conference on imaging systems and techniques (IST). IEEE, pp 510–514
Liu X, Wang C, Bai J, Liao G (2020) Fine-tuning pre-trained convolutional neural networks for gastric precancerous disease classification on magnification narrow-band imaging images. Neurocomputing 392:253–267
Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S (2019) Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 89(357–363):e352
Zhang X, Hu W, Chen F, Liu J, Yang Y, Wang L, Duan H, Si J (2017) Gastric precancerous diseases classification using CNN with a concise model. PLoS ONE 12:e0185508
Wang S, Xing Y, Zhang L, Gao H, Zhang H (2019) Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: experimental feasibility and optimization. Comput Math Methods Med
Wang S, Xing Y, Zhang L, Gao H, Zhang H (2019) A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol 64:235014
Wang S, Cong Y, Zhu H, Chen X, Qu L, Fan H, Zhang Q, Liu M (2020) Multi-scale Context-guided Deep Network for Automated Lesion Segmentation with Endoscopy Images of Gastrointestinal Tract. IEEE J Biomed Health Inf
Krizhevsky A, Hinton G (2009) Learning multiple layers of features from tiny images
Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European conference on computer vision (ECCV), 2018. pp 801–818
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 770–778
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F (2015) WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111
Pogorelov K, Randel KR, de Lange T, Eskeland SL, Griwodz C, Johansen D, Spampinato C, Taschwer M, Lux M, Schmidt PT (2017) Nerthus: a bowel preparation quality video dataset. In: Proceedings of the 8th ACM on multimedia systems conference. pp 170–174
Jha D, Smedsrud PH, Riegler MA, Halvorsen P, de Lange T, Johansen D, Johansen HD (2020) Kvasir-seg: a segmented polyp dataset. In: International conference on multimedia modeling. Springer, pp 451–462
Pogorelov K, Randel KR, Griwodz C, Eskeland SL, de Lange T, Johansen D, Spampinato C, Dang-Nguyen D-T, Lux M, Schmidt PT (2017) Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In: Proceedings of the 8th ACM on multimedia systems conference. pp 164–169
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 3431–3440
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39:2481–2495
Yu F, Koltun V (2015) Multi-scale context aggregation by dilated convolutions. arXiv preprint. arXiv:151107122
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Tang H, Sun N, Li Y (2020) Segmentation model of the opacity regions in the chest X-rays of the Covid-19 patients in the us rural areas and the application to the disease severity. medRxiv
Riaz F, Hassan A, Nisar R, Dinis-Ribeiro M, Coimbra MT (2015) Content-adaptive region-based color texture descriptors for medical images. IEEE J Biomed Health Inf 21:162–171
Fan S, Xu L, Fan Y, Wei K, Li L (2018) Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol 63:165001
Ghatwary N, Ye X, Zolgharni M (2019) Esophageal abnormality detection using densenet based faster r-cnn with gabor features. IEEE Access 7:84374–84385
Lee JH, Kim YJ, Kim YW, Park S, Choi Y-i, Kim YJ, Park DK, Kim KG, Chung J-W (2019) Spotting malignancies from gastric endoscopic images using deep learning. Surg Endosc 33:3790–3797
Khan MA, Khan MA, Ahmed F, Mittal M, Goyal LM, Hemanth DJ, Satapathy SC (2020) Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recogn Lett 131:193–204
Igarashi S, Sasaki Y, Mikami T, Sakuraba H, Fukuda S (2020) Anatomical classification of upper gastrointestinal organs under various image capture conditions using AlexNet. Comput Biol Med 124:103950
Deeba F, Islam M, Bui FM, Wahid KA (2018) Performance assessment of a bleeding detection algorithm for endoscopic video based on classifier fusion method and exhaustive feature selection. Biomed Signal Process Control 40:415–424
e Gonçalves WG, dos Santos MHDP, Lobato FMF, Ribeiro-dos-Santos Â, de Araújo GS (2020) Deep learning in gastric tissue diseases: a systematic review. BMJ Open Gastroenterol 7:e000371
Szczypiński P, Klepaczko A, Pazurek M, Daniel P (2014) Texture and color based image segmentation and pathology detection in capsule endoscopy videos. Comput Methods Progr Biomed 113:396–411
Funding
No funding was received from this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest to report regarding the present study.”
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amin, J., Sharif, M., Gul, E. et al. 3D-semantic segmentation and classification of stomach infections using uncertainty aware deep neural networks. Complex Intell. Syst. 8, 3041–3057 (2022). https://doi.org/10.1007/s40747-021-00328-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00328-7