Abstract
The oral cavity supports a complex and finely balanced consortium of microbial species, many of which cooperate within structured biofilms. These communities develop through multitudinous synergistic and antagonistic interspecies relationships. Changes in the dynamics of oral microbial populations are associated with the transition from healthy teeth and gums to dental caries, gingivitis and periodontitis. Understanding the ecology of oral biofilm communities, and how different species communicate within a given host, will inform new strategies for treatment and prevention of oral diseases. Advances in sequencing technologies have fuelled an increasing trend towards global genomic and proteomic approaches to determine the key factors that initiate oral diseases. Whilst metabolic profiling seeks to identify phenotypic changes of whole microbial communities, transcriptomic studies are exploring their complex interactions with each other and the host. This review discusses the most recent in vitro and in vivo studies of interspecies interactions within polymicrobial oral biofilms.
Similar content being viewed by others
Introduction
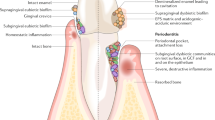
Oral diseases such as dental caries and gingivitis affect 3.9 billion people globally [1]. The involvement of microorganisms in the development of these diseases has long been recognised. Over 700 microbial taxa have been detected in dental plaque, attached to tooth surfaces and in the gingival crevice [2]. In any one individual, 1 gram of mature dental plaque may contain around 109 bacterial cells, belonging to 200 species [3]. These complex microbial communities form highly structured biofilms that develop through a succession of physical and chemical interspecies interactions. Whilst some bacterial species, such as Streptococcus mutans, are major etiological agents of tooth decay [4]; others, such as Porphyromonas gingivalis, have traditionally been more commonly linked to periodontitis [5]. This chronic condition is characterised by inflamed and bleeding gums that lead to bone re-adsorption and tooth loss. Detailed analyses of oral microbial ecology have shown that changes in these species ratios are associated with a shift from health to disease [2, 6].
Recent metagenomic studies, including the human microbiome project, have highlighted further complexity in the relationship between polymicrobial oral communities and human health [3, 7, 8•], and the distinction between “commensal” and “pathogenic” species is no longer clear [9•, 10•]. Advances in experimental model systems and in situ analyses of dental biofilms have identified important interspecies interactions that determine the composition of the oral flora and the interplay between polymicrobial communities and their individual hosts.
This review focuses on recent advances in the study of oral microbial ecology and the interactions between species that govern community development.
Microbial Community Development in the Oral Cavity
Biofilms can be described as communities of microbial cells that are usually attached to a surface, and are protected by an extracellular matrix of secreted polysaccharides, proteins, nucleic acids and lipids [11]. This extracellular polymeric substance (EPS) offers protection, concentration of nutrients and structure to microbial populations [12]. Gradients in pH, O2 and nutrient concentrations have been observed at different biofilm depths [13], creating micro-environments that support the coexistence of diverse microorganisms. Bacterial cells within biofilms have been shown to exhibit slower metabolic rates and increased resistance to antimicrobial agents and immune attack [14, 15]. Significant increases in antibiotic resistance have been demonstrated (4−250-fold) by several oral bacteria when grown as biofilms [16]. The close proximity of microbial cells within biofilms allows for efficient cell−cell signaling; metabolic cooperation and horizontal transfer of genetic material [17].
The striated species composition of mixed biofilm communities in the oral cavity has been reviewed extensively [2, 18–22], and a number of key interacting players have been well described. The specific communities found within each individual host will depend on the outcomes of numerous synergistic and antagonistic relationships, as well as host factors. Oral biofilms develop through a series of successive physical, metabolic and signaling interspecies interactions that have traditionally been grouped into a set of dynamic stages:
Early Birds
Initial colonization of tooth surfaces is mediated by mostly Gram-positive pioneer species, mainly belonging to the genera Streptococci, Veillonella, and Actinomyces [21–26]. Bacterial surface adhesins bind to receptors within the salivary pellicle [18, 20, 22, 27]. Streptococci exhibit wide ranging adhesive properties and metabolic capabilities, enabling them to fill several niches on tooth surfaces and the surrounding soft tissues [23]. Multiplication and growth of these organisms marks the early stages of oral biofilm development. Both synergistic and antagonistic interactions occur between these early arrivals [24–26], to establish successful mixed biofilm communities that alter the environmental conditions in the mouth. In particular, the oral streptococci produce a plethora of general and specific antimicrobial agents in the form of hydrogen peroxide (H2O2), bacteriocins, mucins and lantibiotics in fierce competition for adhesion sites [28, 29]. Conversely, A. naeslundii produces arginine, which is required for growth by oral streptococci [25], and the type II fimbriae of Actinomyces have been shown to bind phosphopolysaccharides on streptococcal surfaces [30]. Host and dietary factors also play a major role in early oral biofilm development. Initial colonization of tooth surfaces can be dramatically altered in the presence of dietary carbohydrates such as sucrose [31].
Regardless of the specific composition of early dental biofilms in the oral cavity of individual hosts, they usually result in reduced pH and oxygen concentrations and provide a new surface, with unique receptor sites, for the attachment of other microbial species [18].
Veillonella species are central to the development of multispecies plaque communities. Whilst they utilise the lactic acid produced by streptococcal species, they produce vitamin K that enhances growth of P. gingivalis, Aggregatibacter actinomycetemcomitans and Prevotella spp [21]. Actimomyces naeslundii and Fusobacterium nucleatum have been termed “bridging species”, as they play a pivotal role in the development of mature dental biofilms [32•]. F. nucleatum can grow in a wide pH range (5.0−7.0) and neutralises acidic environments [33]. A. naeslundii has been shown to co-aggregate with and protect many oral species by consuming H2O2 via protein oxidation [34]. The resultant reduction in oxygen thus creates an permissive environment for the growth of several anaerobes.
Fashionably Late
The late colonisers of oral biofilms are dominated by obligate anaerobes such as P. gingivalis; Tanerella forsythia; Treponema denticola [19, 35] and P. intermedia [33] [36], which produce degradative enzymes and induce inflammatory responses and the destruction of periodontal tissue in vivo. These species have been shown to dramatically increase in numbers in the deep periodontal pockets that characterise advanced periodontitis. Other late colonisers, such as A. actinomycetemcomitans, play a role in the development of localised aggressive periodontitis (LAP) [37].
Cell−cell signalling plays a decisive role in the incorporation of late colonisers into oral biofilm communities. Quorum sensing is a density-dependent signalling system, in which diffusible auto-inducers (AI) regulate the expression of multiple genes. The luxS gene expressing AI-2 is conserved among both Gram-negative and Gram-positive species of oral bacteria, including S. mutans, S. gordonii, S. oralis, P. gingivalis, and A. actinomycetemcomitans [38, 39]. The luxS gene is required for the establishment of mixed biofilms (e.g. between S. oralis and A. naeslundii [18, 40] and between S. gordonii and P. gingivalis [41]). Streptococcal luxS signalling has also been implicated in the formation of fungal Candida albicans biofilms [42].
The synergistic interaction between S. gordonii and P. gingivalis is multifaceted and has been studied extensively. Physical, metabolic and chemical interchanges between the two species have been well characterised. The major fimbriae of P. gingivalis bind to surface-expressed streptococcal glyceraldehyde-3-phosphate dehydrogenase [43], whilst the minor fimbriae bind to SspA/B [44•]. P. gingivalis is asaccharolytic, and does not compete with S. gordonii for sugars. It has been suggested that S. gordonii may deplete oxidants, allowing the survival of the anaerobic P. gingivalis, which secretes several proteases that may breakdown peptides for S. gordonii metabolism [45]. S. gordonii AI-2 is essential for dual species biofilm formation with P. gingivalis in vitro [41] under the control of the transcription factor CdhR [46]. This synergism has also been demonstrated in vivo, leading to increased bone loss in a mouse model of periodontitis [44•].
The New Kids on the Block
Studies of healthy individuals have confirmed that, although each individual mouth harbours unique microbial communities, they share many conserved genera [47]. In addition to Streptococci, and Veillonella spp, Gemella, Granulicatella, Abiotrophia, Selenomonas and Capnocytophaga spp were consistently identified on healthy tooth surfaces [3].
At diseased sites, species diversity has been shown to be more complex and increased numbers of several novel species have been detected. The dominant causative agents of dental caries have traditionally been thought to be mutans streptococci and Lactobacilli. In contrast, Gram-negative anaerobes such as P. gingivalis and T. denticola have been thought of as major etiologic agents of periodontitis. However, newly recognised potential pathogens have triggered a paradigm shift in concepts of pathogenic oral communities [45].
Metagenomic analyses of caries samples suggest a significant role for F. nucleatum, Bifidobacterium species [48] and Scardovia wiggsiae [49]. Studies of subgingival plaque taken during periodontitis have identified potentially pathogenic roles for many more species (e.g. Selenomas spp, Eubacterium spp, Synergistes, Desulfobulbus, TM7, Filifactor alocis [3, 10•, 50, 51]). In light of these discoveries, new theoretical models are moving away from the idea of individual causative agents of oral disease, and towards the suggestion that intricate interactions between oral inhabitants and their specific host may involve a shift in whole community behaviour to become more pathogenic [9•, 10•, 52].
Advances in Model Systems for Studying Oral Biofilms
Despite the recent advances in metagenomic profiling of plaque samples, relevant, simple and well-controlled in vitro models are still required to decipher the physical, spatial, metabolic and chemical basis of these interactions [35, 37]. Dental biofilms have been characterised using in vitro and in vivo models with increasing complexity (reviewed by Palmer et al., 2010 [53]). Table 1 summarises the most recent findings on interspecies interactions in model systems.
The Simplest Models
These involve the growth of static cultures in microtitre plates. Improved biofilm formation on microtitre well surfaces has been achieved using low nutrient concentrations, and hydroxyapatite (the main component of tooth enamel) has been used as a removable substratum. Pre-treatment of surfaces with processed saliva can mimic the salivary pellicle, providing relevant receptors for the adhesion of pioneer species [54]. The multi-well format of these models allows the simultaneous screening of many species combinations for cooperative interactions, the effect of changes in media composition and the comparison of different strains [55].
Polymicrobial biofilms containing A. naeslundii, V. dispar, F. nucleatum, S. sobrinus, and S. oralis have been developed on hydroxyapatite discs [56]. Mixed communities of Streptococcus and Veillonella species, micromanipulated from dental plaque, have been developed using saliva as a sole source of nutrients [57]. More recently, these model systems have been used to identify new mechanisms of cooperation involving protease interactions between T. denticola and P. gingivalis [58, 59•] and AI-2 signalling between S. gordonii and C. albicans [42].
Flow Cell Systems
Such systems involve the continuous flow of media across the model biofilm. They provide adherent cells with replenished nutrients, removing waste products and unattached cells. These systems more closely mimic the environment in the oral cavity and can be used for longer experiments. Palmer et al. (2001) used a flow cell model with 25 % saliva as the sole nutrient source to demonstrate a mutualistic relationship between S. oralis and A. naeslundii, which can only grow as biofilms when cocultured [60]. V. parvula, A. actinomycetemcomitans and F. nucleatum have, more recently, been shown to require co-culture for biofilm growth under flow [37]. By contrast, P. gingivalis, A. oris and V. parvula have been shown to grow in dual-species partnerships, but not as a tri-species community in flow systems.
Adding Complexity
Simple biofilm models comprising two or three species have proved to be an excellent method to determine individual interspecies relationships [35, 58, 59•, 61, 62]. However, many interactions are complex and involve multiple community members. These relationships may not be captured in simple dual or tri-species models. Polymicrobial models could provide a fuller picture of such communities. Bradshaw et al. established a ten-species bioiflm model almost 20 years ago using a stirred vessel reactor [63]. More recently, the ten-species Zurich biofilm model, in microtitre plates, has demonstrated that human serum in the growth medium yields more stable biofilms that are representative of species’ ratios found in periodontitis patients [64•].
Ex-vivo Models
Other biofilm models have been developed by seeding the growth media directly from in vivo oral samples. Biofilms derived from mouse oral microbiota have been shown to inhibit the integration of F. nucleatum upon addition to the system, due to cell contact-dependent induction of hydrogen peroxide [32•]. Others have developed polymicrobial biofilms, using human pooled saliva as the inoculum. The authors reported that 60−80 % of the diversity of the original inoculum was maintained for 48 hours, including many uncultivable species [65].
Considering the Host
The development of oral microbial biofilms and the advancement of periodontal disease are not solely dependent on microbial community dynamics. The in vitro models discussed so far are unable to account for the major contribution of host inflammatory and immune responses [66]. Recent advances in biofilm model systems have included the co-culture of multi-species biofilms with primary gingival epithelial cell lines [67]. Co-culture of a nine-species biofilm with gingival epithelial cells invoked apoptosis, release of pro-inflammatory cytokines and rapid degradation of cytokines. The addition of other relevant host cells, such as neutrophils to these models, will provide further insights into the interaction between polymicrobial biofilms and host factors.
In recent years, a number of polymicrobial infection models have been used to assess the synergistic behaviour of oral microorganisms in vivo. Co-culture of oral streptococci with A. actinomycetemcomitans has been shown to elicit enhanced resistance to killing by host innate immunity in a mouse model [68]. This relationship was also demonstrated to increase pathogenesis in the polymicrobial infection model [69•]. Similarly, simultaneous infection of mice with P. gingivalis and S. gordonii confirmed that interspecies binding was important for P. gingivalis colonization and increased bone loss [44•]. By contract, S. cristatus has been shown to interfere with the pathogenicity P. gingivalis in mice [70•]. In another study, low levels of P. gingivalis have been shown to impair innate immunity and promote the dysbiosis of the symbiotic microbiota, leading to bone loss in vivo. Interestingly, P. gingivalis did not cause periodontitis in germ-free mice. These findings prompted the labelling of P. gingivalis as a “keystone pathogen” [9•]. Synergistic virulence has also been demonstrated between P. gingivalis and T. denticola in a murine periodontitis model [71].
These studies provide evidence that interaction between community members is capable of mediating resistance to host innate immunity and increased virulence, and reinforces the need to understand how polymicrobial growth affects interaction with the host immune system.
Imaging Polymicrobial Oral Communities
Fluorescent in situ hybridisation (FISH) has enabled investigators to visualise snapshots of the spatial arrangements of mixed biofilms either grown within specialised microtitre plates, on removable discs, within flow-cell chambers or in vivo. Similar studies, using a Culture Well chambered coverglass system, have recently been used to determine the physical interaction of the new potential pathogen F. alocis with P. gingivalis, S. gordonii, F. nucleatum and A. actinomycetemcomitans [72]. The three-dimensional (3D) topology of the biofilm structure and detailed spatial arrangements of many interacting species have been visualised [56, 57, 64•, 73–75].
Xiao et al. (2012) use novel imaging techniques to provide insight into the functionality of oral biofilm architecture. They observed how small populations of S. mutans assemble a diffusion-limiting 3D matrix scaffold, which creates localised and highly acidic microenvironments throughout the biofilm. This model could explain the rapid accumulation of cariogenic plaque in the presence of sucrose, even if the initial S. mutans population is numerically and proportionally lower than other cohabitants [76•]
Others have begun to use 3D printing to create bacterial communities that are physically distinct, yet able to communicate through chemical interactions. This approach shows promise for deciphering the importance of spatial arrangements in polymicrobial communities [77].
Developing New Paradigms Through Metagenomics
Over half of the bacterial species identified so far in the human oral cavity are not cultivable in vitro [2]. It is clear, therefore, that classic models based on culture dependent methods can only provide a “drop-in-the-ocean” view of oral microbial communities in vivo (the oral microbiome). Metagenomic approaches are better able to describe the microbial ecology of oral biofilms in situ. Sequencing across highly variable areas of the 16S rRNA gene enables the identification of all the species present in a sample. This approach can provide a comprehensive snap shot of an oral community at a given point in time. DNA microarrays have been used successfully as a high throughput means of screening large numbers of samples for the presence of known species [82]. Further advances in next generation sequencing (NGS) technologies have revolutionised our view of oral microbial communities, and have revealed that overall species diversity may be double than previously thought [45]. As NGS becomes cheaper and faster, it is now possible to perform longitudinal studies of oral biofilm development in large cohorts, providing a more robust picture of the core human oral microbiome.
Recent years have seen an explosion of 16S rRNA sequence data that describe the oral microbiota in samples collected from wide ranging individuals [47, 50, 83–85, 86•]. Reviews of these data have suggested that the progression from health to disease might be more complicated than previously thought [10•, 45]. NGS technologies are also enabling researchers to consider the role of fungi and viruses in oral community stratification [45].
C. albicans is a key player in multiple oral disease conditions, and novel cross-kingdom interactions have been described with streptococcal spp, resulting in increased pathogenicity [42]. A bacterial exoenzyme has been shown to mediate S. mutans interactions with C. albicans by readily attaching to the yeast cell surface. The surface-bound enzyme was found to be functionally active, producing a glucan-rich matrix directly onto the fungal cell wall surface, dramatically enhancing adhesive interactions [87]. Ghannoum et al. (2010) describe the human oral mycobiome from the oral cavity of 20 healthy individuals. Seventy-four culturable and 11 non-culturable fungal genera were common across all samples, with Candida being the most frequently identified, followed by Cladosporium, Aureobasidium, and Saccharomycetales [88•].
Not to be left out, the in vivo viral component of individuals has been termed the human ‘virome’. A robust bacteriophage population has been detected in human saliva [89•] and shared living environments have been suggested to play a significant role in determining the ecology of human oral viruses [90].
Metagenomic studies are providing a picture of oral biofilm communities that is far richer than previously thought. However, these analyses are mainly descriptive and new approaches are required to determine the interactions between these newly identified players.
There is an intricate network of complex interspecies cross-talk in oral microbial communities. Physical ligand-receptor binding, metabolic codependencies, chemical signals that trigger global gene regulators, and chemical warfare combine with host factors to build unique communities in each individual mouth. It has been suggested that many species might be uncultivable because they depend on the secondary byproducts of another microorganism for growth. Deciphering these key signals may identify media supplements that could be used for in vitro culture of these new species [45] or to combat potential pathogens in vivo.
Deep sequencing approaches can now generate whole genome sequences from microbial communities (not just the 16S rRNA gene). This technique has the potential to provide a comprehensive view of global genetic and metabolic changes associated with a given condition. Liu et al. (2012) performed whole metagenome sequencing on subgingival plaque samples from 15 patients. They found that periodontal patient samples were enriched with virulence factors, and detected a common species structure in diseased samples that was not observed in healthy sites. Finally, the depth of sequencing performed in this study enabled the characterisation of the unculturable TM7 genome [8•].
Transcriptomics
Extraction of RNA from biofilm communities and conversion to cDNA enables determination of the levels of expression of specific genes. Real-time, reverse transcription (RT)-PCR and micro-array technologies have enabled the exploration of specific and global transcriptomes (detecting differentially expressed genes in response to a given stimulus). Investigation of the transcriptional responses of P. gingivalis to growth within an oral bacterial community detected that 18 % of the genome was differentially expressed compared to growth in monocultures [91]. Cell envelope biogenesis, DNA replication, energy production and fatty acid metabolism were downregulated, indicating a decrease in growth and replication. In contrast, transport systems, adhesins, signal transduction and transcriptional regulation were upregulated, indicating multi-level interspecies interactions [91].
There have been several transcriptomic studies of the partnership between S. gordonii and P. gingivalis. Differentially expressed genes in the dual-species environment have been mainly involved in metabolism and energy production, but also in potential virulence factors such as a tyrosine phosphatase [19, 46]. Similar studies have demonstrated that co-culture with T. denticola promoted increased expression of P. gingivalis adhesin genes and proteases [59•]. In contrast, a negative interaction was found between P. gingivalis and S. cristatus that downregulated expression of P. gingivalis fimbrial (fimA) genes, and interfered with biofilm formation [92].
Transcriptomic characterisation of a polymicrobial biofilm model has recently been reported, using microarray chips that include the genomes of five oral species (S. sanguinis, S. mutans, F. nucleatum, A. actinomycetemcomitans and P. gingivalis [78]. It is now possible to compare global gene expression in more than one species in response to one another’s presence (the “interactomes” of mixed microbial communities) using RNASeq. Frias-Lopez et al. (2011) used this technique for metatranscriptomic analysis of a multispecies biofilm model (A. naeslundii, L. casei, S. mitis, V. parvula, and F. nucleatum). Addition of P. gingivalis and A. actinomycetemcomitans to the system induced upregulation of chaperones, ABC transport systems and putative transposases. Alterations in the expression of small RNAs provided evidence of interspecies gene expression control [93•].
In addition, Murine GeneChip(R) microarrays [94] have been used to determine the host response to polymicrobial infections (P. gingivalis, T. denticola, and T. forsythia) using a murine calvarial model of acute inflammation and bone resorption [95•].
Proteomics
Proteomic approaches are now being applied to the investigation of interspecies interactions using simple, well-controlled, in vitro model systems. The technique is able to identify and quantify all of the peptides within a given sample by mass spectrometry (2D capillary HPLC/MS/MS analysis). Peptide sequences are processed and concatenated using a range of sequence databases to identify each protein. The relative abundance of each protein can also be calculated to gain quantitative information about each interaction. By comparing the protein content of a single species community with that of a mixed culture, it is now possible to identify all of the proteins that are differentially expressed in response to a second or third microbial population [61, 96•, 97•, 98]. Ontology analysis tools, such as DAVID [99, 100], can infer meaning to the lists of differentially expressed proteins by clustering them according to function in specific pathways.
This method has been used to compare the envelope proteins of P. gingivalis cultures with those that were co-cultured with an oral bacterial community [101]. Twenty-four surface proteins, including RgpA, were upregulated during biofilm growth and 18 proteins were downregulated. When compared to transcriptional profiles of the same system [91], correlation between differentially expressed mRNA and proteins was marginal, reflecting the multi-faceted regulatory networks that control bacterial responses to complex environments [19].
Whole cell quantitative proteomics have recently been applied to models of community interaction between S. gordonii, F. nucleatum and P. gingivalis (Sg-Fn-Pg) [96•, 98], and between P. gingivalis, T. denticola and T. forsythia (Pg-Td-Tf) [79]. These studies reported extensive changes in P. gingivalis protein expression in response to the other species. In the Sg-Fn-Pg model, the cell envelope proteome and vitamin synthesis pathways were affected [98]. The S. gordonii proteome exhibited increased energy metabolism, amino acid biosynthesis, exopolysaccharide and lactate production, and oxidative stress protection mechanisms in response to the mixed environment. A decrease in sugar transport, adhesion and ethanol production was observed. Several interactions were found to be species specific or more prominent in response to one species compared to another. Acetate levels were decreased in response to F. nucleatum, but increased in response to P. gingivalis, whilst formate was downregulated only in the presence of P. gingivalis [96•]. In the absence of exogenous nutrients, these proteomic profiles are consistent with S. gordonii utilising nutrients produced by the other community members and downregulating adhesins after initial contact. Analysis of mixed Pg-Td-Tf biofilm communities in flow cell systems identified differential expression of proteins involved in iron acquisition, nutrient sharing, flagella structure and the secretion of virulence factors.
A high-throughput quantitative proteomics approach has also been used to examine how S. mutans interacts within mixed-species biofilms. Klein et al. (2012) used state-of-the-art MudPIT technology to describe the proteomic profile of S. mutans when cultured in biofilms with A. naeslundii and S. oralis. This study also quantified the abundance of S. mutans proteins produced in mixed species environments in the presence of sucrose. Proteins involved in exopolysaccharide matrix assembly, metabolic and stress adaptation processes were shown to increase as the biofilms matured. Specific adaptation mechanisms were also upregulated as an adaptive response to acidic environments. Fundamental differences in matrix assembly were observed in mixed versus single species environment [97•].
The proteomics studies discussed here demonstrate the scale of interspecies interactions between common inhabitants of the oral cavity and the intricate physiological processes that are linked to expression of virulence within complex biofilms. Mixed communities showed significant changes in 45−54 % of the detected proteome compared to single species controls [96•]. This highlights the relatively little we still know about the intricacies of communication within oral microbial communities. The proteomic approach has potential to identify a wide range of previously unrecognised mechanisms of interaction. As technologies and pathway deciphering tools become more sophisticated, these analyses will be able to elucidate these mechanisms in more complex communities.
Metabolomics
Metabolomics is the comprehensive identification and quantification of all the metabolites present in a given biological sample at a given time. Most metabolites are small, polar, ionic molecules (e.g. amino acids, nucleotides, carboxylic acids, phosphorylated sugars) and can be separated using chromatography or, more recently, capillary electrophoresis (CE), then identified and quantified using high-resolution mass spectrometry (MS) [81]. This technique can determine the activity of many important metabolic pathways. In particular, sugar metabolism in plaque samples is of interest to researchers in the field of oral microbiology. Furthermore, metabolic profiles of bacterial co-culture supernatants can identify mechanisms of bacteria−bacteria interaction by metabolite exchange. This novel approach has the capacity to reveal global and dynamic biological information, and can be used to comprehensively analyse intracellular and extracellular metabolites. Takahashi et al. (2010) compared the metabolic profiles of supragingival plaque samples from healthy volunteers with pure cultures of oral bacterial species S. mutans, S. sanguinis, A. oris, and A. naeslundii, before and after glucose treatment. They showed that the EMP pathway, pentose-phosphate pathway and TCA cycle were active both in supra gingival plaque taken directly from healthy patients and in pure cultures of representative plaque bacteria (Streptococcus spp and Actimomyces spp). Metabolic profiles were shown to change considerably following treatment with glucose. These data validate the uses of metabolomic profiling on in vitro model systems to characterise interspecies interactions in dental plaque. Data obtained from metabolomic studies have been suggested to reflect phenotypic changes more closely than proteomics or transcriptomics. It is interesting to note that, while large numbers of oral pathogen genomes have been sequenced, little quantitative information is known about their metabolic functions. Mazumdar et al. (2009) report the computational construction of a genome-scale metabolic network for P. gingivalis, including 679 metabolic reactions. The network enables prediction of the phenotypic effects of gene knockouts in given conditions. Predictive computer modelling will provide clues towards the successful culture of new potential pathogens, and may be able to map novel interspecies interactions.
Conclusions
The new oral microbiology research summarised in this review illustrates an evolution of interspecies interaction studies, from heavily culture-based investigations to use of new high-throughput sequencing technologies. Metagenomic studies have further elucidated the immense diversity of oral microbial ecology. They have inspired new dogma that suggests a role for community, rather than individual species, in the delicate balance between health and disease [10•, 52]. These data have, so far, been mainly descriptive and have generated unfathomable sequence information, a large portion of which is assigned to unnamed and uncultivable microorganisms. The clinical implications of these large sequence data sets are still to be fully resolved. The mere presence or numerical superiority may not necessarily be linked with the pathogenesis of the disease. Deeper sequencing, along with transcriptomic and proteomic analysis of clinical samples could highlight changes in the behaviour of these microbial communities and their interplay with the host. Future directions include more systems biology approaches that have developed new bioinformatic tools for assigning function to genes [80, 99, 102]. These enable biosynthetic pathways of newly recognised species to be determined, and allow researchers to predict the behaviour of specific communities. Such information will provide clues about the pathogenicity of given communities. Identification of crucial biosynthetic pathways will instruct us as to how previously uncultivable species might be cultured and any novel therapeutic potential that they might harbour. In the same way, systems level analysis and molecular imaging of complex ex vivo communities [103] will inform state-of-the-art modelling of the spatial and temporal relationships between oral biofilm members at different physical niches within the mouth.
Computational analysis promises to add meaning to big data sets by combining metagenomic, transcriptomic, proteomic, and metabolomic studies, and building pathways to investigate global processes within whole communities [80, 104]. At the same time, it is important not to forget the power of small, simple in vitro model systems that can illustrate the mechanisms of individual interspecies interactions that are relevant in vivo with elegance and class.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Marcenes W et al. Global burden of oral conditions in 1990–2010: a systematic analysis. J Dent Res. 2013;92(7):592–7.
Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13(12):589–95.
Dewhirst FE et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–17.
Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90(3):294–303.
Socransky SS et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–44.
Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–90.
Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18(2):109–20.
Liu B et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7(6):e37919. This article demonstrates the use of deep whole metagenomic sequencing to better characterise the oral microbiota in sub-gingival plaque samples. The authors report the genome sequence of the unclultivable potential pathogenTM7. The study exemplifies how next generation sequencing can be used to determine important biosynthetic pathways for clues on how to culture newly identified species, and the exploitable resources that they may provide.
Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–25. This review article introduces the keystone pathogen hypothesis that low abundance microbial pathogens can remodel the healthy microbiota to trigger inflammatory disease.
Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–19. This review article introduces the conceptual theory of polymicrobial synergy and disbiosis model (PSD), in which periodontitis is initiated by a synergistic and disbiotic microbial community, rather than a select few periopathogens. The authors summarise a strong body of evidence from recent research to support the model. The article represents a major shift in how oral microbial communities are interpreted in health and disease.
Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–33.
Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2(5):435–44.
Vroom JM et al. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl Environ Microbiol. 1999;65(8):3502–11.
Hoiby N et al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–32.
Harriott MM, Noverr MC. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother. 2010;54(9):3746–55.
Sedlacek MJ, Walker C. Antibiotic resistance in an in vitro subgingival biofilm model. Oral Microbiol Immunol. 2007;22(5):333–9.
Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8(12):1441–50.
Kolenbrander PE et al. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–80.
Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52(1):38–52.
Wright CJ et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28(2):83–101.
Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192(12):2965–72.
Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3(2):49–54.
Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73(3):407–50. Table of Contents.
Jakubovics NS et al. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66(3):637–44.
Jakubovics NS et al. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008;190(10):3646–57.
Palmer Jr RJ et al. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol. 2003;185(11):3400–9.
Nobbs AH, Jenkinson HF, Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J Dent Res. 2011;90(11):1271–8.
Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28(8):397–403.
Wen ZT et al. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 2010;10:111.
Yang J et al. Comparative structural and molecular characterization of ribitol-5-phosphate-containing Streptococcus oralis coaggregation receptor polysaccharides. J Bacteriol. 2009;191(6):1891–900.
Schilling KM, Bowen WH. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun. 1992;60(1):284–95.
He X et al. Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb Ecol. 2012;63(3):532–42. This article describes an in vitro biofilm model, developed by seeding the growth media directly from in vivo oral samples. F. nucleatum was prevented from integrating into the biofilms derived from mouse oral microbiota due to cell contact-dependent induction of hydrogen peroxide.
Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 2003;18(2):109–13.
Periasamy S et al. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl Environ Microbiol. 2009;75(10):3250–7.
Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191(22):6804–11.
van Winkelhoff AJ et al. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29(11):1023–8.
Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. 2009;77(9):3542–51.
Bassler BL. Small talk. Cell-to-cell communication in bacteria. Cell. 2002;109(4):421–4.
Xie H et al. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. 2007;153(Pt 10):3228–34.
Rickard AH et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60(6):1446–56.
McNab R et al. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185(1):274–84.
Bamford CV et al. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77(9):3696–704.
Nagata H et al. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun. 2009;77(11):5130–8.
Daep CA et al. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79(1):67–74. This article reports that the increased bone loss observed on simultaneous infection of mice with P. gingivalis and S. gordonii is dependent on dual-species adhesion. This confirms that interspecies interactions have an effect in vivo.
Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13(12):3077–87.
Chawla A et al. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. 2010;78(6):1510–22.
Bik EM et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4(8):962–74.
Ventura M et al. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009;5(12):e1000785.
Tanner AC et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49(4):1464–74.
Griffen AL et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–85.
Kumar PS et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44(10):3665–73.
Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81(2):305–14.
Palmer, R.J., Jr., Supragingival and subgingival plaque: paradigm of biofilms. Compend Contin Educ Dent, 2010. 31(2):104–6, 108, 110 passim; quiz 124, 138.
Lemos JA et al. Protocols to study the physiology of oral biofilms. Methods Mol Biol. 2010;666:87–102.
Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8(5):607–20.
Guggenheim M et al. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl Environ Microbiol. 2001;67(3):1343–50.
Chalmers NI et al. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190(24):8145–54.
Cogoni V et al. Treponema denticola chymotrypsin-like proteinase (CTLP) integrates spirochaetes within oral microbial communities. Microbiology. 2012;158(Pt 3):759–70.
Meuric V et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28(1):40–53. This article reports that T. denticola causes upregulation of P. gingivalis haemagglutinin binding proteins and proteases.
Palmer Jr RJ et al. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69(9):5794–804.
Kuboniwa M et al. Insights into the virulence of oral biofilms: discoveries from proteomics. Expert Rev Proteomics. 2012;9(3):311–23.
Kaplan CW et al. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for interspecies adherence and the structured architecture of multispecies biofilm. Mol Microbiol. 2009;71(1):35–47.
Bradshaw DJ, Marsh PD. Effect of sugar alcohols on the composition and metabolism of a mixed culture of oral bacteria grown in a chemostat. Caries Res. 1994;28(4):251–6.
Ammann TW, Gmur R, Thurnheer T. Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol. 2012;12:227. This article reports the improvement of a ten-species biofilm model by the addition of human serum to yield more stable biofilms with species ratios that are representative of those found in periodontitis patients.
Edlund A et al. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 2013;1(1):25.
Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 2013;62(1):203–17.
Guggenheim B et al. In vitro modeling of host-parasite interactions: the 'subgingival' biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009;9:280.
Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106(5):1578–83.
Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7(3):e1002012. This report uses a mouse model to demonstrate that metabollic interactions between oral streptococci and A. actinomycetemcomitans lead to increase virulence in vivo. This is important, because it shows that interactions identified in in vitro models are relevant in the presence of host factors.
Xie H et al. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. 2012;47(5):578–83. This article reports the use a mouse model to demonstrate that the antagonistic interaction between S. cristatus and P. gingivalis interferes with the pathogenicity of P. gingivalis in vivo.
Orth RK et al. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011;26(4):229–40.
Wang Q et al. Oral Community Interactions of Filifactor alocis In Vitro. PLoS One. 2013;8(10):e76271.
Kuboniwa M et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60(1):121–39.
Diaz PI et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72(4):2837–48.
Zijnge V et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5(2):e9321.
Xiao J et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. This study illustrates how the EPS-matrix and microcolonies are assembled tri-dimensionally, while simultaneously mapping the pH of intact mixed-species biofilms in 3D using novel imaging techniques.
Connell, J.L., et al., 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A, 2013
Redanz S et al. A five-species transcriptome array for oral mixed-biofilm studies. PLoS One. 2011;6(12):e27827.
Zainal-Abidin Z et al. Differential proteomic analysis of a polymicrobial biofilm. J Proteome Res. 2012;11(9):4449–64.
Mazumdar V et al. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191(1):74–90.
Takahashi N, Washio J, Mayanagi G. Metabolomics of supragingival plaque and oral bacteria. J Dent Res. 2010;89(12):1383–8.
Paster BJ, Dewhirst FE. Molecular microbial diagnosis. Periodontol 2000. 2009;51:38–44.
Xie G et al. Community and gene composition of a human dental plaque microbiota obtained by metagenomic sequencing. Mol Oral Microbiol. 2010;25(6):391–405.
Colombo AP et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80(9):1421–32.
Griffen AL et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6(4):e19051.
Abusleme L et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7(5):1016–25. This article exemplifies many of the recent oral microbiome studies that associate certain microbial species with disease symptoms.
Gregoire S et al. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 2011;77(18):6357–67.
Ghannoum MA et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6(1):e1000713. This first report characterises the fungal microbiota of the oral cavity, and will pave the way for more investigation into the role of more fungal species in oral disease.
Pride DT et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6(5):915–26. This article demonstrates the ability of next generation sequencing technologies to describe the viral component of the oral cavity in health and disease, and the role that viruses might play in oral community stratification.
Robles-Sikisaka R et al. Association between living environment and human oral viral ecology. ISME J. 2013;7(9):1710–24.
Lo AW et al. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18.
Wang BY et al. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47(12):3902–6.
Frias-Lopez J, Duran-Pinedo A. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol. 2012;194(8):2082–95. This article is one of the first to compare global gene expression in more than one oral microbial species in response to the mixed species environment using RNASeq.
Handfield M, Baker HV, Lamont RJ. Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87(3):203–23.
Bakthavatchalu V et al. Polymicrobial periodontal pathogen transcriptomes in calvarial bone and soft tissue. Mol Oral Microbiol. 2011;26(5):303–20. This key paper describes the host transcriptional response to polymicrobial infections (P. gingivalis, T. denticola, and T. forsythia) using a murine calvarial model of acute inflammation and bone resorption.
Hendrickson EL et al. Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol. 2012;12. This article reports the use of whole cell quantitative proteomics to investigate global changes in protein expression in S. gordonii in response to a mixed-species environment containing P. gingivalis and F. nucleatum. The data show that energy metabolism pathways are generally upregulated whilst sugar transport is downregulated in the mixed environment, but that interactions are species specific. The study validates the proteomics approach for investigating interspecies interactions and highlights the importance of nutrient transfer for community interaction.
Klein MI et al. Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics. PLoS One. 2012;7(9):e45795. The authors of this article used the state-of-the-art MudPIT technology, a high-throughput large-scale quantitative proteomic method, to assess both the proteomic profile and protein abundance within mixed-species oral biofilms.
Kuboniwa M et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98.
da Huang W et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183.
Hendrickson EL, Lamont RJ, Hackett M. Tools for interpreting large-scale protein profiling in microbiology. J Dent Res. 2008;87(11):1004–15.
Ang CS et al. Application of 16O/18O reverse proteolytic labeling to determine the effect of biofilm culture on the cell envelope proteome of Porphyromonas gingivalis W50. Proteomics. 2008;8(8):1645–60.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Valm AM et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108(10):4152–7.
Dimitrov DV, Hoeng J. Systems approaches to computational modeling of the oral microbiome. Front Physiol. 2013;4:172.
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
Dr. Chloe James declares no potential conflicts of interest relevant to this article.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
James, C.E. Recent Advances in Studies of Polymicrobial Interactions in Oral Biofilms. Curr Oral Health Rep 1, 59–69 (2014). https://doi.org/10.1007/s40496-013-0006-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40496-013-0006-3