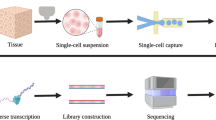
Abstract
Neural stem cells (NSCs) in the adult central nervous system play essential roles in both normal homeostasis and repair of damaged tissue after injury. The study of adult NSCs is hampered by the heterogeneous NSC population. In this review, we describe recent progresses in using single-cell RNA-sequencing (scRNA-seq) technique for the investigation of NSCs. The first part of this review focuses on the scRNA-seq techniques and bioinformatic analysis. The second part emphasizes the applications of scRNA-seq analysis in NSC research. Finally, we discuss the challenges and future directions of scRNA-seq technique for both basic research and regenerative medicine.

Similar content being viewed by others
References
Achim K, Pettit JB, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33(5):503–9. doi:10.1038/nbt.3209.
Adams DL, Zhu P, Makarova OV, Martin SS, Charpentier M, Chumsri S, et al. The systematic study of circulating tumor cell isolation using lithographic microfilters. RSC Adv. 2014;9:4334–42. doi:10.1039/C3RA46839A.
Alvarez-Buylla A, Kohwi M, Nguyen TM, Merkle FT. The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harb Symp Quant Biol. 2008;73:357–65. doi:10.1101/sqb.2008.73.019.
Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7(6):744–58. doi:10.1016/j.stem.2010.11.017.
Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–25. doi:10.1016/j.cell.2014.04.005.
Bifari F, Decimo I, Pino A, Llorens-Bobadilla E, Zhao S, Lange C, et al. Neurogenic radial glia-like cells in meninges migrate and differentiate into functionally integrated neurons in the neonatal cortex. Cell Stem Cell. 2016; doi:10.1016/j.stem.2016.10.020.
Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17(4):385–95. doi:10.1016/j.stem.2015.09.003.
Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10(11):1093–5. doi:10.1038/nmeth.2645.
Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112(51):15672–7. doi:10.1073/pnas.1520760112.
Choi JH, Ogunniyi AO, Du M, Du M, Kretschmann M, Eberhardt J, et al. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol Prog. 2010;26(3):888–95. doi:10.1002/btpr.374.
DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–2. doi:10.1093/bioinformatics/bts196.
Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci. 2010;11(5):339–50. doi:10.1038/nrn2822.
Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6(11):1127–34. doi:10.1038/nn1144.
du Verle DA, Yotsukura S, Nomura S, Aburatani H, Tsuda K. CellTree: an R/bioconductor package to infer the hierarchical structure of cell populations from single-cell RNA-seq data. BMC Bioinformatics. 2016;17(1):363. doi:10.1186/s12859-016-1175-6.
Dulin JN, Antunes-Martins A, Chandran V, Costigan M, Lerch JK, Willis DE, et al. Transcriptomic approaches to neural repair. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35(41):13860–7. doi:10.1523/JNEUROSCI.2599-15.2015.
Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586–603. doi:10.1038/nprot.2006.85.
Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–8.
Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8(6):469–77. doi:10.1038/nmeth.1613.
Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17(3):175–88. doi:10.1038/nrg.2015.16.
Grun D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163(4):799–810. doi:10.1016/j.cell.2015.10.039.
Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, et al. Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Scientific reports. 2016;6:31851. doi:10.1038/srep31851.
Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi:10.1186/s13059-016-0888-1.
Ishii Y, Matsumoto Y, Watanabe R, Elmi M, Fujimori T, Nissen J, et al. Characterization of neuroprogenitor cells expressing the PDGF beta-receptor within the subventricular zone of postnatal mice. Mol Cell Neurosci. 2008;37(3):507–18. doi:10.1016/j.mcn.2007.11.006.
Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–6. doi:10.1038/nmeth.2772.
Juliá M, Telenti A, Rausell A. Sincell: an R/Bioconductor package for statistical assessment of cell-state hierarchies from single-cell RNA-seq. Bioinformatics. 2015;31(20):3380–2. doi:10.1093/bioinformatics/btv368.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi:10.1186/1471-2105-9-559.
Leng N, Chu L-F, Barry C, Li Y, Choi J, Li X, et al. Oscope identifies oscillatory genes in unsynchronized single cell RNA-seq experiments. Nat Methods. 2015;12(10):947–50. doi:10.1038/nmeth.3549.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. doi:10.1093/bioinformatics/btp698.
Linnarsson S. Sequencing single cells reveals sequential stem cell states. Cell Stem Cell. 2015;17(3):251–2. doi:10.1016/j.stem.2015.08.016.
Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7(3):179–93. doi:10.1038/nrn1867.
Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17(3):329–40. doi:10.1016/j.stem.2015.07.002.
Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161(5):1175–86. doi:10.1016/j.cell.2015.04.001.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14. doi:10.1016/j.cell.2015.05.002.
Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70(4):687–702. doi:10.1016/j.neuron.2011.05.001.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi:10.1038/nature09807.
Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18(5):591–6. doi:10.1016/j.stem.2016.03.012.
Petersen E, Wilson ME, Touch S, McCloskey B, Mwaba P, Bates M, et al. Rapid spread of Zika virus in the Americas—implications for public health preparedness for mass gatherings at the 2016 Brazil Olympic games. Int J Infect Dis. 2016;44:11–5. doi:10.1016/j.ijid.2016.02.001.
Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–8. doi:10.1038/nmeth.2639.
Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–81. doi:10.1038/nprot.2014.006.
Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16(2):85–97. doi:10.1038/nrg3868.
Ross HH, Ambrosio F, Trumbower RD, Reier PJ, Behrman AL, Wolf SL. Neural stem cell therapy and rehabilitation in the central nervous system: emerging partnerships. Phys Ther. 2016;96(5):734–42. doi:10.2522/ptj.20150063.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi:10.1038/nbt.3192.
Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17(3):360–72. doi:10.1016/j.stem.2015.07.013.
Sun J, Sun J, Ming GL, Song H. Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur J Neurosci. 2011;33(6):1087–93. doi:10.1111/j.1460-9568.2011.07607.x.
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi:10.1038/nmeth.1315.
Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18(5):587–90. doi:10.1016/j.stem.2016.02.016.
Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–13. doi:10.1038/nature20123.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. doi:10.1038/nbt.2859.
Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534(7607):391–5. doi:10.1038/nature18323.
Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58(4):598–609. doi:10.1016/j.molcel.2015.05.005.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–5. doi:10.1093/bioinformatics/bts356.
Welberg L. Learning and memory: neurogenesis erases existing memories. Nat Rev Neurosci. 2014;15(7):428. doi:10.1038/nrn3768.
Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, et al. Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from Zika virus infection. Cell Stem Cell. 2016;19(6):703–8. doi:10.1016/j.stem.2016.11.011.
Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–7. doi:10.1038/nature12364.
Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011;192(3):373–382. doi:10.1083/jcb.201010021.
Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30(8):777–82.
Acknowledgments
The work was supported in part by grants from the New Jersey Commission on Spinal Cord Research and Busch Biomedical Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there is no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the topical collection on Bioinformatics and Stem Cell
Rights and permissions
About this article
Cite this article
Li, Y., Anderson, J., Kwan, K.Y. et al. Single-Cell Transcriptome Analysis of Neural Stem Cells. Curr Pharmacol Rep 3, 68–76 (2017). https://doi.org/10.1007/s40495-017-0084-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40495-017-0084-3