Abstract
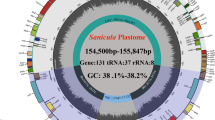
The genus Smilax (Smilacaceae), commonly known as Sarsaparilla, comprises about 262 species with numerous medicinal and economic importance. Due to considerable morphological similarity, Smilax has been recognized as a taxonomically challenging group. In this study, we conducted a comprehensive analysis of the genomic architecture and nucleotide variation within the genus Smilax, comparing the newly sequenced plastome of Smilax zeylanica with ten other plastomes. Our analyses revealed a highly conserved gene structure, order, and orientation across the plastomes studied. Nonetheless, we identified eight highly divergent regions, namely rbcL-accD, petA-psbJ, psaJ-rpl33, ndhC-trnV UAC, accD-psaI, ndhF-rpl32, trnK UUU, and rps16-trnQ UUG. These highly diverse DNA regions could potentially be used as DNA super-barcodes for the precise identification of Smilax species. Furthermore, our study identified four positively selected genes—accD, matK, psaA, and rbcL. We also observed the loss of infA and pseudogenization of ycf15 and ycf68 genes within Smilacaceae. Additionally, the prediction of RNA editing sites revealed a high level of conservation across the genus Smilax. These findings provide valuable insights into adaptation, evolutionary dynamics, marker development, and barcode validation in Smilax, ultimately enhancing its therapeutic applications.







Similar content being viewed by others
Data availability
All data generated or analysed during the current study are included in this published article, NCBI repository (Accession No. OL701493) [and its supplementary information files].
References
Abdala S, Martin-Herrera D, Benjumea D, Perez-Paz P (2008) Diuretic activity of Smilax canariensis, an endemic Canary Island species. J Ethnopharmacol 119:12–16. https://doi.org/10.1016/j.jep.2005.03.005
Amiryousefi A, Hyvönen J, Poczai P (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34:3030–3031. https://doi.org/10.1093/bioinformatics/bty220
Andrews S (2010) Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc
Anonymous (1950) The wealth of India, Vol. VIII. New Delhi, Council of scientific and industrial research, pp. 365
Arriola ÍA, Melo-Junior JC, Ferreira BG, Isaias R (2018) Galls on Smilax campestris Griseb. (Smilacaceae) protect the insects against restinga constraints, but do not provide enriched nutrition. Rev Bras Bot 41:145–153. https://doi.org/10.1007/s40415-017-0423-y
Bánki O, Roskov Y, Döring M, Ower G et al. (2023) Catalogue of life checklist (Version 2023–10–19). Catalogue of life. https://doi.org/10.48580/dft7
Barthelson RA, Sundareshan P, Galbraith DW, Woosley RL (2006) Development of a comprehensive detection method for medicinal and toxic plant species. Am J Bot 93:566–574. https://doi.org/10.3732/ajb.93.4.566
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585. https://doi.org/10.1093/bioinformatics/btx198
Bock DG, Andrew RL, Rieseberg LH (2014) On the adaptive value of cytoplasmic genomes in plants. Mol Ecol 23:4899–4911. https://doi.org/10.1111/mec.12920
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Boudreau E, Turmel M (1995) Gene rearrangements in Chlamydomonas chloroplast DNAs are accounted for by inversions and by the expansion/contraction of the inverted repeat. Plant Mol Biol 27:351–364. https://doi.org/10.1007/BF00020189
Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S, Comparative Sequencing Program NISC (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731. https://doi.org/10.1101/gr.926603
Bullard AJ, Allen CM (2013) Synopsis of the woody species of Smilax in the eastern United States north of peninsular Florida. J North Carolina Acad Sci 129:37–43. https://doi.org/10.7572/2167-5880-129.2.37
Challinor VL, Parsons PG, Chap S, White EF, Blanchfield JT, Lehmann RP, De Voss JJ (2012) Steroidal saponins from the roots of Smilax sp.: structure and bioactivity. Steroids 77:504–511. https://doi.org/10.1007/s13659-017-0139-5
Chan PP, Lowe TM (2019) tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Kollmar M (ed) Gene prediction. Humana, New York, pp 1–14. https://doi.org/10.1007/978-1-4939-9173-0_1
Chase MW, Fay MF, Devey DS, Maurin O, Rønsted N, Davies TJ, Pillon Y, Peterson G, Tamura MN, Asmussen CB, Hilu K (2006) Multigene analyses of monocot relationships. Aliso A J Syst Florist Botany 22:63–75
Chen D, Zhao Y, Zhao D (1993) Identification study of the Chinese drug Tu fuling and its adulterants. CJ PA 13:302–306
Chen SC, Qiu YX, Wang AL, Cameron KM, Fu CX (2006) A phylogenetic analysis of the Smilacaceae based on morphological data. Act Phytotax Sin 44:113–125. https://doi.org/10.1360/aps050065
Commision CP (2015) The pharmacopoeia of the people’s republic of China, 2015th edn. China medical science press, China, pp 496–498
D’Agostino N, Tamburino R, Cantarella C, De Carluccio V, Sannino L, Cozzolino S, Cardi T, Scotti N (2018) The complete plastome sequences of eleven Capsicum genotypes: insights into DNA variation and molecular evolution. Genes 9:503. https://doi.org/10.3390/genes9100503
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18. https://doi.org/10.1093/nar/gkw955
Do HD, Kim JS, Kim JH (2013) Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes (Melanthiaceae). Gene 530:229–235. https://doi.org/10.1016/j.gene.2013.07.100
Do HD, Kim C, Chase MW, Kim JH (2020) Implications of plastome evolution in the true lilies (monocot order Liliales). Mol Phylogenet Evol 148:106818. https://doi.org/10.1016/j.ympev.2020.106818
Dong JL, Sun BN, Li AJ, Chen H (2021) The diversity of Smilax (Smilacaceae) leaves from the middle miocene in southeastern China. Geol J 56:744–757. https://doi.org/10.1002/gj.3882
Donoghue MJ, Baldwin BG, Li J, Winkworth RC (2004) Viburnum phylogeny based on chloroplast trnK intron and nuclear ribosomal ITS DNA sequences. Syst Bot 29:188–198. https://doi.org/10.1600/036364404772974095
Evans WC (2007) Trease and evans pharmacognosy, 15th edn. Saunders Company, New Delhi, p 476
Fay MF, Chase MW, Rønsted N, Devey DS, Pillon Y, Pires JC, Peterson G, Seberg O, Davis JI (2006) Phylogenetics of Liliales. Aliso A J Syst Florist Botany 22:559–565. https://doi.org/10.5642/aliso.20062201.43
Feng JY, Jin XJ, Zhang SL, Yang JW, Fei SP, Huang YS, Feng JY, Jin XJ, Zhang SL, Yang JW, Fei SP, Huang YS, Liu Y, Li QZC, P, (2022) Smilax weniae, a new species of smilacaceae from limestone areas bordering Guizhou and Guangxi. China Plants 11:1032. https://doi.org/10.3390/plants11081032
Ferrufino-Acosta L (2014) Taxonomic revision of the genus Smilax (Smilacaceae) in central America and the Caribbean Islands. Willdenowia 40:227–280. https://doi.org/10.3372/wi.40.40208
Floden A, Schilling EE (2018) Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Mol Phylogenet Evol 129:202–213. https://doi.org/10.1016/j.ympev.2018.08.017
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279. https://doi.org/10.1093/nar/gkh458
Gao F, Chen C, Arab DA, Du Z, He Y, Ho SY (2019) EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol 9:3891–3898. https://doi.org/10.1002/ece3.5015
Givnish TJ, Zuluaga A, Marques I, Lam VK, Gomez MS, Iles WJ, Ames M, Spalink D, Moeller JR, Briggs BG, Lyon SP (2016) Phylogenomics and historical biogeography of the monocot order Liliales: out of Australia and through Antarctica. Cladistics 32:581–605. https://doi.org/10.1186/s12915-021-01166-2
Gomes Pacheco T, de Santana LA, de Oliveira JD, Campos Otoni W, Balsanelli E, de Oliveira PF, Maltempi de Souza E, Rogalski M (2020) The complete plastome of Passiflora cirrhiflora A. Juss.: structural features, RNA editing sites, hotspots of nucleotide diversity and molecular markers within the subgenus Deidamioides. Rev Bras Bot 43:839–853. https://doi.org/10.1007/s40415-020-00655-y
Harpke D, Peruzzi L, Kerndorff H, Karamplianis T, Constantinidis T, Randelovic V, Randelovic N, Juskovic M, Pasche E, Blattner FR (2014) Phylogeny, geographic distribution, and new taxonomic circumscription of the Crocus reticulatus species group (Iridaceae). Turk J Bot 38:1182–1198. https://doi.org/10.3906/bot-1405-60
Hausner G, Olson R, Simon D, Johnson I, Sanders ER, Karol KG, McCourt RM, Zimmerly S (2006) Origin and evolution of the chloroplast trnK (matK) intron: a model for evolution of group II intron RNA structures. Mol Biol Evol 23:380–391. https://doi.org/10.1093/molbev/msj047
Howe CJ, Barbrook AC, Koumandou VL, Nisbet RE, Symington HA, Wightman TF (2003) Evolution of the chloroplast genome. Philos Trans R Soc Lond B Biol Sci 358:99–107. https://doi.org/10.1098/rstb.2002.1176
Hu C, Zheng X, Teng Y (2011) Characterization and phylogenetic utility of non-coding chloroplast regions trnL-trnF and accD-psaI in Pyrus. Acta Hortic Sin 38:2261–2272
Hu S, Sablok G, Wang B, Qu D, Barbaro E, Viola R, Li M, Varotto C (2015) Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genom 16:1–4. https://doi.org/10.1186/s12864-015-1498-0
Hu Y, Zhang Z, Wu B, Gao J, Li Y (2016) Genetic relationships of buckwheat species based on the sequence analysis of ITS and ndhF-rpl32. Biodivers Sci 24:296. https://doi.org/10.17520/biods.2015259
Ichinose M, Sugita M (2016) RNA editing and its molecular mechanism in plant organelles. Genes 8:5. https://doi.org/10.3390/genes8010005
Jain SK, Rao RR (1977) Handbook of field and herbarium methods. Today and Tomorrows Publishers, New Delhi
Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. PNAS 104:19369–19374
Jena PK, Dinda SC, Ellaiah P (2012) Phytochemical investigation and simultaneous study on antipyretic, anticonvulsant activity of different leafy extracts of Smilax zeylanica Linn. Orient Pharm Exp Med 12:123–127. https://doi.org/10.1007/s13596-011-0052-0
Jyothi T, Rabinanaryan A, Shukla CP, Harisha CR (2013) Phytochemical studies on Smilax macrophylla linn. A source plant of Chopacheeni. Global J Res Med Plants Indigen Med 2:110
Katoh K, Toh H (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. https://doi.org/10.1093/bioinformatics/btq224
Kesanakurti P, Thirugnanasambandam A, Ragupathy S, Newmaster SG (2020) Genome skimming and NMR chemical fingerprinting provide quality assurance biotechnology to validate Sarsaparilla identity and purity. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-76073-7
Kim JS, Kim JH (2013) Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea). PLoS ONE 8:e68180. https://doi.org/10.1371/journal.pone.0068180
Kim KJ, Lee HL (2004) Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res 11:247–261. https://doi.org/10.1093/dnares/11.4.247
Kim JS, Hong JK, Chase MW, Fay MF, Kim JH (2013) Familial relationships of the monocot order Liliales based on a molecular phylogenetic analysis using four plastid loci: matK, rbcL, atpB and atpF-H. Bot J Linn 172:5–21. https://doi.org/10.1371/journal.pone.0068180
Kirtikar KR, Basu BD (1991) Indian medicinal plants. In: Dehra D, Bishen S, Mahendra Pal S, eds, pp. 2496
Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44:237–244. https://doi.org/10.1111/j.1365-313X.2005.02533.x
Kritpetcharat O, Khemtonglang N, Kritpetcharat P et al (2011) Using DNA markers and barcoding to solve the common problem of identifying dried medicinal plants with the examples of Smilax and Cissus in Thailand. J Med Plant Res 5:3480–3487
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642. https://doi.org/10.1093/nar/29.22.4633
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. https://doi.org/10.1093/nar/gkh152
Lenz H, Hein A, Knoop V (2018) Plant organelle RNA editing and its specificity factors: enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinform 19:1–8. https://doi.org/10.1186/s12859-018-2244-9
Liu Q, Xue Q (2005) Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J Genet 84:55–62. https://doi.org/10.1007/BF02715890
Liu J, Qi ZC, Zhao YP, Fu CX, Xiang QY (2012) Complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales–influences of gene partitions and taxon sampling. Mol Phylogenet Evol 64:545–562. https://doi.org/10.1016/j.ympev.2012.05.010
Lu RS, Yang T, Chen Y, Wang SY, Cai MQ, Cameron KM, Li P, Fu CX (2021) Comparative plastome genomics and phylogenetic analyses of Liliaceae. Bot J Linn Soc 196:279–293. https://doi.org/10.1093/botlinnean/boaa109
Madoka Y, Tomizawa KI, Mizoi J, Nishida I, Nagano Y, Sasaki Y (2002) Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol 43:1518–1525. https://doi.org/10.1093/pcp/pcf172
Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, Kolganova TV, Skyabin KG (2008) Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol 66:555–564. https://doi.org/10.1007/s00239-008-9091-7
Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165. https://doi.org/10.1038/30234
Martins AR, Soares MK, Redher VL, Bajay MM, Villela P, Zucchi MI, Appezzato-da-Glória B (2014) Use of anatomical, chemical, and molecular genetic characteristics in the quality control of medicinal species: a case study of Sarsaparilla (Smilax spp.). Econ Bot 68:410–425. https://doi.org/10.1007/s12231-014-9287-2
McNeal JR, Leebens-Mack JH, Arumuganathan K, Kuehl JV, Boore JL, DePamphilis CW (2006) Using partial genomic fosmid libraries for sequencing complete organellar genomes. Biotechniques 41:69–73. https://doi.org/10.2144/000112202
Michelangeli FA, Guimaraes PJ, Penneys DS, Almeda F, Kriebel R (2013) Phylogenetic relationships and distribution of new world Melastomeae (Melastomataceae). Bot J Linn Soc 171:38–60. https://doi.org/10.1111/j.1095-8339.2012.01295.x
Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13:645–658. https://doi.org/10.1105/tpc.13.3.645
API (2011) The Ayurvedic Pharmacopoeia of India (Part I, volume V). New Delhi: Ministry of Health and Family Welfare, Government of India
Ohsako T, Fukuoka S, Bimb HP, Baniya BK, Yasui Y, Ohnishi O (2001) Phylogenetic analysis of the genus Fagopyrum (Polygonaceae), including the Nepali species F. megacarpum, based on nucleotide sequence of the rbcL-accD region in chloroplast DNA. Fagopyrum 18:9–14
Oomachan MM, Masih SK (1991) Ethno-botanical and conservation aspects of medicinal plants of Madhya Pradesh. J Pure and Appli Sci 6:39
Palmer JD (1991) Plastid chromosomes: structure and evolution. In: Indra V (ed) The molecular biology of plastids, 1st edn. Academic press, pp 5–53
Park I, Yang S, Kim WJ, Noh P, Lee HO, Moon BC (2018) Authentication of herbal medicines Dipsacus asper and Phlomoides umbrosa using DNA barcodes, chloroplast genome, and sequence characterized amplified region (SCAR) marker. Molecules 23:1748. https://doi.org/10.3390/molecules23071748
Patel S, Biswas B, Rambabu K, Sundaram E, Arya R (2020) Pharmacognostical studies of Smilax aspera Linn. A herbal drug. Indian J Res Homoeopathy 14:260–260. https://doi.org/10.4103/ijrh.ijrh_13_20
Pereram BPR (2014) A study on the plants used as Chopachini. J Homeop Ayurv Med 3:2167–1206. https://doi.org/10.4172/2167-1206.1000170
Pharmacopoeia commission of the ministry of health of the people’s republic of China (2020) Pharmacopoeia of the people’s republic of China, 1st edn. China Medical Science Press, Beijing, China
Piot A, Hackel J, Christin PA, Besnard G (2018) One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 247:255–266. https://doi.org/10.1007/s00425-017-2781-x
POWO (2023) Plants of the world online. Facilitated by the royal botanic gardens, Kew. Published on the Internet; http://www.plantsoftheworldonline.org/ Accessed 10 Oct 2023
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. https://doi.org/10.1371/journal.pone.0009490
Qi Z, Cameron KM, Li P, Zhao Y, Chen S, Chen G, Fu C (2013) Phylogenetics, character evolution, and distribution patterns of the greenbriers, Smilacaceae (Liliales), a near-cosmopolitan family of monocots. Bot J Linn 173:535–548. https://doi.org/10.1111/boj.12096
Rajesh V, Perumal P (2014) In vivo assessment of antidiabetic and antioxidant activities of methanol extract of Smilax zeylanica leaves in wistar rats. Orient Pharm Exp Med 14:127–144. https://doi.org/10.1007/s13596-013-0137-z
Raman G, Park S, Lee EM, Park S (2019) Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-41377-w
Raubeson LA, Jansen RK (2005) Chloroplast genomes of plants. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. CABI, UK, pp 45–68. https://doi.org/10.1079/9780851999043.0045
Raúl SC, Beatriz HC, Joseoziel LG, Francenia SSN (2017) Phenolic compounds in genus Smilax (Sarsaparilla). Phenolic compounds: natural sources, importance, and applications, 1st edn. Intech Open Book, p 233
Resorts (2017) Ayurvedic medicinal plants of Sri Lanka. 2017. http://www.instituteofayurveda.org/plants
Rocha MJ, Batista JA, Guimarães PJ, Michelangeli FA (2016) Phylogenetic relationships in the Marcetia alliance (Melastomeae, Melastomataceae) and implications for generic circumscription. Bot J Linn Soc 181:585–609. https://doi.org/10.1111/boj.12429
Rockenbach K, Havird JC, Monroe JG, Triant DA, Taylor DR, Sloan DB (2016) Positive selection in rapidly evolving plastid–nuclear enzyme complexes. Genetics 204:1507–1522. https://doi.org/10.1534/genetics.116.188268
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Ruhlman TA, Jansen RK (2014) The plastid genomes of flowering plants. In: Maliga P (ed) Chloroplast biotechnology. Humana Press, Totowa, pp 3–38. https://doi.org/10.1007/978-1-62703-995-6_1
Selvaraj D, Sarma RK, Sathishkumar R (2008) Phylogenetic analysis of chloroplast matK gene from Zingiberaceae for plant DNA barcoding. Bioinformation 3:24–27. https://doi.org/10.6026/97320630003024
Shahrajabian MH, Sun W, Cheng Q (2019) Tremendous health benefits and clinical aspects of Smilax china. Afr J Pharm Pharmacol 13:253–258. https://doi.org/10.5897/AJPP2019.5070
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166. https://doi.org/10.3732/ajb.92.1.142
Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB (2014) Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot 101:1987–2004. https://doi.org/10.3732/ajb.1400398
Sofiah S, Sulistyaningsih ld, (2019) The diversity of Smilax (Smilacaceae) in Besiq-Bermai and bontang forests, east Kalimantan, Indonesia. Biodiversitas 20:379–387
Soledispa PA, González J, Cuéllar A, Pérez J, Monan M (2018) GC-MS chemical characterization of main components of Smilax Domingensis wild in Cuba. J Agric Res 6:79–86. https://doi.org/10.5296/jas.v6i2.13099
Srirama R, Santhosh Kumar JU, Seethapathy GS, Newmaster SG, Ragupathy S, Ganeshaiah KN, Uma Shaanker R, Ravikanth G (2017) Species adulteration in the herbal trade: causes, consequences and mitigation. Drug Saf 40:651–661. https://doi.org/10.1007/s40264-017-0527-0
Srivastava JG (1971) The botanical identity of ‘Vacha’(‘Bachh’) of the ayurvedic literature. Q J Crude Drug Res 11:1734–1742
Steele PR, Friar LM, Gilbert LE, Jansen RK (2010a) Molecular systematics of the neotropical genus Psiguria (Cucurbitaceae): implications for phylogeny and species identification. Am J Bot 97:156–173. https://doi.org/10.3732/ajb.0900192
Steele KP, Ickert-Bond SM, Zarre S, Wojciechowski MF (2010b) Phylogeny and character evolution in Medicago (Leguminosae): evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences. Am J Bot 97:1142–1155. https://doi.org/10.3732/ajb.1000009
Takenaka M, Zehrmann A, Verbitskiy D, Haertel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47:335–352. https://doi.org/10.1146/annurev-genet-111212-133519
Tang Y, He X, Quanlan C, Lanlan F, Jianye Z, Zhongzhen Z, Tang Y, He X, Quanlan C, Lanlan F, Jianye Z, Zhongzhen Z, Dong L, Zhitao L, Yi T, Chen H (2014) A mixed microscopic method for differentiating seven species of “Bixie”-related Chinese materia medica. Microsc Res Tech 77:57–70. https://doi.org/10.1002/jemt.22313
Techaprasan J, Klinbunga S, Ngamriabsakul C, Jenjittikul T (2010) Genetic variation of Kaempferia (Zingiberaceae) in Thailand based on chloroplast DNA (psbA-trnH and petA-psbJ) sequences. Genet Mol Res 9:1957–1973. https://doi.org/10.4238/vol9-4gmr873
Techen N, Crockett SL, Khan IA, Scheffler BE (2004) Authentication of medicinal plants using molecular biology techniques to compliment conventional methods. Curr Med Chem 11:1391–1401. https://doi.org/10.2174/0929867043365206
Tian LW, Zhang Z, Long HL, Zhang YJ (2017) Steroidal saponins from the genus Smilax and their biological activities. Nat Prod Bioprospecting 7:283–298. https://doi.org/10.1007/s13659-017-0139-5
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6-11. https://doi.org/10.1093/nar/gkx391
Tsutsumi C, Zhang XC, Kato M (2008) Molecular phylogeny of Davalliaceae and implications for generic classification. Syst Bot 33:44–48. https://doi.org/10.12705/656.2
Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM (2008) Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol 8:1–14. https://doi.org/10.1186/1471-2148-8-36
Wang M, Bai QX, Zheng XX, Hu WJ, Wang S, Tang HP, Yu AQ, Yang BY, Kuang HX (2023) Smilax china L.: a review of its botany, ethnopharmacology, phytochemistry, pharmacological activities, actual and potential applications. J Ethnopharmaco. https://doi.org/10.1016/j.jep.2023.116992
Wicke S, Schneeweiss GM, Depamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297. https://doi.org/10.1007/s11103-011-9762-4
Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM (2013) Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. https://doi.org/10.1105/tpc.113.113373
Wilson TC, Conn BJ, Henwood MJ (2012) Molecular phylogeny and systematics of Prostanthera (Lamiaceae). Aust Syst Bot 25:341–352. https://doi.org/10.1071/SB12006
Wong KH, Siu TY, Tsang SS, Kong BL, Wu HY, But GW, Hui JH, Shaw PC, Lau DT (2023) The complete chloroplast genomes of nine Smilacaceae species from Hong Kong: Inferring infra-and inter-familial phylogeny. Int J Mol Sci 24(8):7460. https://doi.org/10.3390/ijms24087460
Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, Daniell H, Duvall MR, Lin CS (2010) Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol 10:1–12. https://doi.org/10.1186/1471-2229-10-68
Wu JY, Liu YSC, Ding ST, Li J, An PC (2017) Late Pliocene Smilax (Smilacaceae) leaves from Southwest China: phytogeographical and paleoecological implications. Rev Palaeobot Palynol 241:26–38. https://doi.org/10.1016/j.revpalbo.2017.02.006
Yang T, Liao X, Yang L, Liu Y, Mu W, Sahu SK, Liu X, Strube ML, Zhong B, Liu H (2019) Comparative analyses of 3654 chloroplast genomes unraveled new insights into the evolutionary mechanism of green plants. bioRxiv 655241. https://doi.org/10.1101/655241
Ye WQ, Yap ZY, Li P, Comes HP, Qiu YX (2018) Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae). Mol Phylogenet Evol 127:978–987. https://doi.org/10.1016/j.ympev.2018.07.001
Yi Y, Cao Z, Yang D, Cao Y, Wu Y, Zhao S (1998) Studies on the chemical constituents of Smilax glabra. Yao Xue Xue Bao Acta Pharmaceutica Sinica 33(11):873–5. https://doi.org/10.3390/molecules18055265
Yu X, Tan W, Gao H, Miao L, Tian X (2020) Development of a specific mini-barcode from plastome and its application for qualitative and quantitative identification of processed herbal products using DNA metabarcoding technique: a case study on Senna. Front Pharmacol 11:585687. https://doi.org/10.3389/fphar.2020.585687
Zhang W, Sun Y, Liu J, Xu C, Zou X, Chen X, Liu Y, Wu P, Yang X, Zhou S (2021) DNA barcoding of Oryza: conventional, specific, and super barcodes. Plant Mol Biol 105:215–228. https://doi.org/10.1007/s11103-020-01054-3
Zhang Y, Zhang L, Lu Y, He X, Zhang H (2023) Characterization of the complete mitochondrial genome and phylogenetic analysis of Epiverta chelonia (Coleoptera: Coccinellidae). Mitochondrial DNA B 8:7–9. https://doi.org/10.1080/23802359.2022.2157196
Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A (2020) Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet 11:1123. https://doi.org/10.3389/fgene.2020.576124
Zhu ZM, Gao XF, Fougère-Danezan M (2015) Phylogeny of Rosa sections Chinenses and Synstylae (Rosaceae) based on chloroplast and nuclear markers. Mol Phylogenet Evol 87:50–64. https://doi.org/10.1016/j.ympev.2015.03.014
Acknowledgements
The authors thank the Director of Agharkar Research Institute for encouragement and facilities. Financial support received from Science and Engineering Research Board-Department of Science and Technology (SERB-DST) vide Project no. CRG/2020/000910 to carry out this research is duly acknowledged. Partial support received by the Department of Science & Technology, New Delhi, to GS via Inspire fellowship vide ref. no. IF210309 is also acknowledged. We also thank Prof. M. M. Sardesai and Dr. M.N. Datar for their help in the sample collection.
Funding
This work was supported by the Science and Engineering Research Board-Department of Science and Technology (SERB-DST) vide Project no. CRG/2020/000910. Partial support was received from the Department of Science & Technology, New Delhi, to GS via Inspire fellowship vide ref. no. IF210309.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study's conception and design. GS and SM curated the data and performed the analysis. GS drafted the initial manuscript. RKC and SM reviewed and revised the manuscript. RKC acquired funding for this research. All authors read and approved the final manuscript as submitted.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sukhramani, G., Maurya, S. & Choudhary, R.K. Plastome comparison reveals hotspots of nucleotide diversity and positive selection pressure on accD, matK, psaA and rbcL genes in Smilacaceae. Braz. J. Bot 47, 145–161 (2024). https://doi.org/10.1007/s40415-023-00973-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-023-00973-x