Abstract
Introduction
Detection of adverse reactions to drugs and biologic agents is an important component of regulatory approval and post-market safety evaluation. Real-world data, including insurance claims and electronic health records data, are increasingly used for the evaluation of potential safety outcomes; however, there are different types of data elements available within these data resources, impacting the development and performance of computable phenotypes for the identification of adverse events (AEs) associated with a given therapy.
Objective
To evaluate the utility of different types of data elements to the performance of computable phenotypes for AEs.
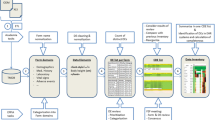
Methods
We used intravenous immunoglobulin (IVIG) as a model therapeutic agent and conducted a single-center, retrospective study of 3897 individuals who had at least one IVIG administration between 1 January 2014 and 31 December 2019. We identified the potential occurrence of four different AEs, including two proximal AEs (anaphylaxis and heart rate alterations) and two distal AEs (thrombosis and hemolysis). We considered three different computable phenotypes: (1) an International Classification of Disease (ICD)-based phenotype; (2) a phenotype-based on EHR-derived contextual information based on structured data elements, including laboratory values, medication administrations, or vital signs; and (3) a compound phenotype that required both an ICD code for the AE in combination with additional EHR-derived structured data elements. We evaluated the performance of each of these computable phenotypes compared with chart review-based identification of AEs, assessing the positive predictive value (PPV), specificity, and estimated sensitivity of each computable phenotype method.
Results
Compound computable phenotypes had a high positive predictive value for acute AEs such as anaphylaxis and bradycardia or tachycardia; however, few patients had both ICD codes and the relevant contextual data, which decreased the sensitivity of these computable phenotypes. In contrast, computable phenotypes for distal AEs (i.e., thrombotic events or hemolysis) frequently had ICD codes for these conditions in the absence of an AE due to a prior history of such events, suggesting that patient medical history of AEs negatively impacted the PPV of computable phenotypes based on ICD codes.
Conclusions
These data provide evidence for the utility of different structured data elements in computable phenotypes for AEs. Such computable phenotypes can be used across different data sources for the detection of infusion-related adverse events.
Similar content being viewed by others
References
United States Food and Drug Adminitration. What is a Serious Adverse Event? [Internet]. 2022. https://www.fda.gov/safety/reporting-serious-problems-fda/what-serious-adverse-event. Accessed 10 Jan 2022.
Coloma PM, Trifirò G, Patadia V, Sturkenboom M. Postmarketing safety surveillance: where does signal detection using electronic healthcare records fit into the big picture? Drug Saf. 2013;36:183–97.
Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013;20:e226–31.
Hurst JH, Liu Y, Maxson PJ, Permar SR, Boulware LE, Goldstein BA. Development of an electronic health records datamart to support clinical and population health research. J Clin Transl Sci. 2020;5: e13.
Tang M, Goldstein BA, He J, Hurst JH, Lang JE. Performance of a computable phenotype for pediatric asthma using the problem list. Ann Allergy Asthma Immunol. 2020;125:611-613.e1.
Graham J, Iverson A, Monteiro J, Weiner K, Southall K, Schiller K, et al. Applying computable phenotypes within a common data model to identify heart failure patients for an implantable cardiac device registry. Int J Cardiol Heart Vasc. 2022;39: 100974.
Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi J-C, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43:1130–9.
Weeks J, Pardee R. Learning to share health care data: a brief timeline of influential common data models and distributed health data networks in U.S. Health Care Research. EGEMS (Wash DC). 2019;7:4.
Bonilla FA. Intravenous and subcutaneous immunoglobulin G replacement therapy. Allergy Asthma Proc. 2016;37:426–31.
Perez EE, Orange JS, Bonilla F, Chinen J, Chinn IK, Dorsey M, et al. Update on the use of immunoglobulin in human disease: a review of evidence. J Allergy Clin Immunol. 2017;139:S1-46.
Guo Y, Tian X, Wang X, Xiao Z. adverse effects of immunoglobulin therapy. Front Immunol. 2018;9:1299.
Ammann EM, Haskins CB, Fillman KM, Ritter RL, Gu X, Winiecki SK, et al. Intravenous immune globulin and thromboembolic adverse events: a systematic review and meta-analysis of RCTs. Am J Hematol. 2016;91:594–605.
Cherin P, Marie I, Michallet M, Pelus E, Dantal J, Crave J-C, et al. Management of adverse events in the treatment of patients with immunoglobulin therapy: a review of evidence. Autoimmun Rev. 2016;15:71–81.
Späth PJ, Granata G, La Marra F, Kuijpers TW, Quinti I. On the dark side of therapies with immunoglobulin concentrates: the adverse events. Front Immunol. 2015;6:11.
R Core Team. R: A language and environment for statistical computing. [Internet]. R Foundation for Statistical Computing. http://www.R-project.org/. Accessed 10 Jan 2022.
Di Giovanni R, Cochrane A, Parker J, Lewis DJ. Adverse events in the digital age and where to find them. Pharmacoepidemiol Drug Saf. 2022;31(11):1131–1139. https://doi.org/10.1002/pds.5532
Markvardsen LH, Christiansen I, Harbo T, Jakobsen J. Hemolytic anemia following high dose intravenous immunoglobulin in patients with chronic neurological disorders. Eur J Neurol. 2014;21:147–52.
Shin H, Lee S. An OMOP-CDM based pharmacovigilance data-processing pipeline (PDP) providing active surveillance for ADR signal detection from real-world data sources. BMC Med Inform Decis Mak. 2021;21:159.
Martinez C, Wallenhorst C, van Nunen S. Intravenous immunoglobulin and the current risk of moderate and severe anaphylactic events, a cohort study. Clin Exp Immunol. 2021;206:384–94.
Spadaro G, Vultaggio A, Alberto Bosi A, Reichert D, Janssen J, Lamacchia D, et al. Rapid infusions of human normal immunoglobulin 50g/l are safe and well tolerated in immunodeficiencies and immune thrombocytopenia. Int Immunopharmacol. 2017;44:38–42.
Arnold DM, Heddle NM, Cook RJ, Hsia C, Blostein M, Jamula E, et al. Perioperative oral eltrombopag versus intravenous immunoglobulin in patients with immune thrombocytopenia: a non-inferiority, multicentre, randomised trial. Lancet Haematol. 2020;7:e640–8.
Savaşan S, Tuzcu V, Warrier I, Karpawich P. Cardiac rhythm abnormalities during intravenous immunoglobulin G infusion for treatment of thrombocytopenia. J Pediatr Hematol Oncol. 1997;19:254–7.
Jin PH, Shin SC, Dhamoon MS. Risk of thrombotic events after inpatient intravenous immunoglobulin or plasma exchange for neurologic disease: A case-crossover study. Muscle Nerve. 2020;62:327–32.
Wittstock M, Benecke R, Zettl UK. Therapy with intravenous immunoglobulins: complications and side-effects. Eur Neurol. 2003;50:172–5.
Ammann EM, Cuker A, Carnahan RM, Perepu US, Winiecki SK, Schweizer ML, et al. Chart validation of inpatient International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) administrative diagnosis codes for venous thromboembolism (VTE) among intravenous immune globulin (IGIV) users in the Sentinel Distributed Database. Medicine (Baltimore). 2018;97: e9960.
Bruggeman CW, Nagelkerke SQ, Lau W, Manlhiot C, de Haas M, van Bruggen R, et al. Treatment-associated hemolysis in Kawasaki disease: association with blood-group antibody titers in IVIG products. Blood Adv. 2020;4:3416–26.
Scott DE, Epstein JS. Hemolytic adverse events with immune globulin products: product factors and patient risks. Transfusion. 2015;55:S2-5.
Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23:1046–52.
PCORnet: The National Patient-Centered Clinical Research Network [Internet]. [cited 2018 Oct 27]. https://pcornet.org/. Accessed 10 Jan 2022.
Welcome to OMOP – OHDSI [Internet]. [cited 2022 Apr 12]. https://ohdsi.org/omop/. Accessed 10 Jan 2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the US Food and Drug Administration (FDA) (75F40120C00123 to BAG). This publication reflects the views of the authors and should not be construed to represent FDA's views or policies.
Conflicts of Interest
None declared.
Ethics Approval
This study was declared exempt from review by the DUHS IRB (protocol # Pro00104993).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
Data will be made available by request to the authors.
Code Availability
Code will be made available upon request to the authors.
Author Contributions
JHH, AB, CZ, CKZ, HW, and BAG conceived and planned the analyses. AB and CZ extracted the data and conducted the analyses. JHH drafted the manuscript. JHH, HD, HPH, MP, BR, MDS, IS, MER, and JJS conducted the chart review. MDS, IS, MER, JJS, and GMD provided key clinical expertise to inform the development of the computable phenotypes. All authors provided critical feedback and helped shape the research, analysis, and manuscript. All authors read and approved the final version of the manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hurst, J.H., Brucker, A., Zhao, C. et al. Use of Structured Electronic Health Records Data Elements for the Development of Computable Phenotypes to Identify Potential Adverse Events Associated with Intravenous Immunoglobulin Infusion. Drug Saf 46, 309–318 (2023). https://doi.org/10.1007/s40264-023-01276-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-023-01276-6