Abstract
A microstructure analytics and 3D reconstruction software package, DREAM.3D, was integrated as a module into a cloud-based platform, BisQue. Parallelization of DREAM.3D module executions and the ability to parameterize pipeline variables over a range of values have led to insights about the grain segmentation misorientation tolerance in TriBeam-collected 3D EBSD datasets of additively manufactured materials with complex anisotropic microstructures. Furthermore, a comparison in grain size measurements was made between standard 2D metallographic slices and 3D measures using BisQue’s parallelized DREAM.3D module executions. The direction of cloud-based data infrastructure and the prospects for impact in material science are also discussed.
Similar content being viewed by others
Introduction
Recent external pressures mandated by government grants requiring data be accessible to the public, and internal pressures within academic communities for data sharing and reproducibility have led to the emergence of data-driven platforms within the scientific community [1,2,3,4]. Requirements for these platforms include providing data provenance, data sharing, and hosting services for data that may otherwise disappear when a lab closes or hardware fails [5]. Recently, data repositories have shifted from simply siloing data to include data analytics and predictive capabilities. Furthermore, some of these repositories are directly linked with high-performance computing in order to populate an ever growing database, such as with thermodynamic databases such as the Materials Project [6] and the Open Quantum Materials Database (OQMD) [7]. More sophisticated predictive/interactive databases, such as the aforementioned thermodynamic ones, have been slower to develop with the 3D Materials Science community. This is likely due to the less standardized and more diverse data modalities, the complexity and diversity of microstructures, and the relative infancy of the community itself.
In almost all regards, using 3D data to answer scientific questions in Materials Science is challenging due to the need for dataset acquisition, reconstruction, and analysis. Reconstruction of 3D datasets is a complex process with many degrees of freedom. The algorithms employed, and tolerances selected by the user to align slices, segment features, and remove data artifacts (i.e., remove noise), ultimately determine the quantitative data extracted from the volume. Significant progress on the reconstruction and analysis of electron backscatter diffraction (EBSD) data has been made by a software package DREAM.3D [8], which creates a unified data structure for 3D data treatment and aggregates tools and algorithms collectively developed by the community. In DREAM.3D, this process takes the form of a pipeline, which typically consists of well over a dozen steps, and in which each step has its own set of input variables and outputs. DREAM.3D typically runs in a serial process on a workstation, with predefined input parameters. Due to the long computation times required to process 3D data, which often scales proportionately as dataset size and complexity increases, it is impractical to explore the full parameter space by manually running reconstructions. Typically, only several variables are adjusted by a user until a qualitatively acceptable reconstruction is attained. This process inherently imparts bias and typically provides little opportunity to justify specific reconstruction variables.
The bias generated during reconstruction of 3D datasets is subsequently carried through to data analysis, which, for microstructural data, is still an ill-defined process. Standardization of analysis involving new characterization techniques is generally a slow process. Automated orientation imaging analysis via EBSD first emerged in the early 90s, but standards did not emerge for nearly two decades [9, 10]. These new standards covered basic quantitative analysis, including methods for grain size measurement, as well as guidelines governing data quality, quantity, and resolution sufficient to make such measurements [11]. Even in 2D EBSD analysis, such standards are rare and do not necessarily address additional subtleties arising from more complex, albeit commonly studied, microstructures, including twinned materials, nanocrystalline materials, and deformed materials [12,13,14,15,16,17]. Standardization of characterization methods becomes critical as the complexity of the analyses increases. For example, high-resolution EBSD (HR-EBSD) and Heaviside Digital Image Correlation (HDIC) [18] are extensions of conventional EBSD and digital image correlation (DIC) that require specialized post-processing to measure elastic and plastic strain at a resolution of 10-4 or smaller [19, 20] and 1.5×10− 3 [21], respectively. The variety of analytic approaches available to make accurate strain measurements, as well as the sensitivity of these measurements to instrumentation and sample preparation, motivates more rigorous standardization of the technique via sample reference standards [22, 23].
In 3D characterization techniques, such as X-ray-based diffraction contrast tomography (DCT), the problems of data reconstruction and analysis are also convoluted. Coherent volumes must be reconstructed from individual 2D images via back projection techniques [24]. The diffraction spots from the 2D projections are generally assumed to originate from a single grain, limiting the characterization of materials with large internal strain gradients [25]. Moreover, stereological conventions established in 2D do not necessarily capture the real 3D microstructures, particularly at extrema of property distributions [26, 27]. Given the wealth of additional data provided by 3D characterization, new tools aimed at exploring the parameter space offered to 3D reconstruction are required. In the following manuscript, we describe the use of a cloud-based platform, BisQue [28, 29], enhanced with the DREAM.3D microstructural software analysis package for the 3D reconstruction and analysis of additively manufactured microstructures characterized using TriBeam tomography [30].
Methods
The following sections describe the software infrastructure and materials investigated.
BisQue for Cloud-Based Data Storage and Analysis
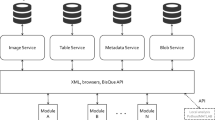
All data processing was performed in the cloud with BisQue [28, 29] using a web browser interface to interact with the 3D datasets. BisQue has backend infrastructure support for web services such as Amazon S3, IRODS, and federated local file systems as well as client facing tools such as Java, Python, and Matlab. A DREAM.3D [8] plugin was developed for BisQue in order to incorporate the materials science based 3D dataset analysis tools. The database architecture of BisQue is such that every processing execution, dataset (input and output), and workflow processing pipeline receives a unique web address URL. These URLs are useful for at least two reasons: tracking data provenance and data sharing. Data provenance with BisQue is automatically captured and can be easily accessed through visualized workflows, as shown in Fig. 1. The workflow shows where input data is located, the parameters of a module execution are captured in the metadata, and the output data location is tracked. Therefore, passing data between research colleagues becomes straightforward due to the data operation transparency. Furthermore, data sharing is facilitated due to the data all being stored and accessed remotely, mitigating versioning problems common to large data operations. For example, two groups collaborating between the US and Europe may find difficulty operating on the same dataset version, especially when business hours do not overlap, without extensive communication.
BisQue DREAM.3D Integration
A DREAM.3D module was incorporated into BisQue to allow for the processing and treatment of 3D datasets with the advantages of cloud-based infrastructure described in the previous section (“BisQue for Cloud Based Data Storage and Analysis”). The DREAM.3D processing capabilities were also extended to allow for parallelized processing across wide parameter spaces. Anytime a processing pipeline is generated in DREAM.3D through BisQue, parameter input variables can be represented as a range of values (with sub-increments), rather than as a distinct value. An example of DREAM.3D pipeline execution is shown in Fig. 2 alongside the analogous representation in DREAM.3D. The user selects an input source and pipeline source, parameterizable values are then automatically displayed in the parameter section. Using straightforward list comprehension, many independent instantiations of DREAM.3D can be run at once across multiple nodes on a compute cluster, dramatically decreasing setup and analysis time for detailed parameter studies. The resulting output files can then be examined as individual pipelines as BisQue has a fully integrated table viewer that allows for both table viewing and inspection of the tree structure of HDF files. In addition, a volume viewer enables web-based visualization of cell-level data, removing the need for programs like HDFView or ParaView and their associated hardware requirements [31, 32]. 3D volumes collected via TriBeam tomography and reconstructed in DREAM.3D generally require many tens of gigabytes of RAM for visualization in ParaView, limiting analysis to powerful workstation computers.
DREAM.3D pipeline for reconstruction of an additively manufactured 304L Stainless Steel 3D dataset as viewed in (a) DREAM.3D on a workstation computer and (b) the DREAM.3D BisQue module in a web browser. The DREAM.3D BisQue module allows for parameterization for variable inputs in a DREAM.3D pipeline, which can be executed in parallel on high-performance compute clusters and then analyzed to determine the ideal reconstruction parameters
Materials and Data Collection
The 3D datasets were collected via TriBeam tomography from two additively manufactured alloys in their as-built state, meaning no additional heat treatments were performed on the samples after additive processing. TriBeam tomography [33] utilizes a femtosecond laser for micromachining within a dual-beam scanning electron microscope (SEM) equipped with a focused ion beam (FIB). The ultrashort pulse length minimizes damage to the machined surface while enabling high material removal rates, roughly four to five orders of magnitude faster than FIB milling alone, resulting in an accessible volume on the order of a cubic millimeter [30, 34]. The FIB can also be used at a glancing angle to minimize surface roughness due to laser-induced periodic surface structures (LIPSS); however, this cleaning step is material dependent and is not always required to obtain high-quality 3D EBSD data.
LENS-Processed 304L Stainless Steel
Laser engineered net shaping (LENS) is a directed energy deposition (DED) additive manufacturing technique derived from laser cladding processes, in which metal powder is blown into a molten pool created using a laser. The low dilution area and high deposition rates achievable in LENS, up to an order of magnitude higher than powder-bed additive processes, allow for the manufacture of large scale parts [35]. A sample of 304L stainless steel fabricated by the LENS process was characterized in the TriBeam system using a slice thickness of 3 μm. The femtosecond laser ablated surface was cleaned using glancing-angle FIB milling to increase the quality of EBSD patterns. EBSD scans were collected on each slice with an in-plane resolution of 3 μm, yielding square voxels. Roughly 200 GB of data was collected from a total of 80 slices over the course of two days, resulting in a volume of 816 × 834 × 240μm. Raw EBSD patterns, comprising nearly 90% of the collected data, were saved for post-processing. Individual slices were reconstructed into a coherent volume using DREAM.3D in BisQue. A minimum confidence index (CI) of 0.1 was chosen as the threshold for data quality, as the indexing of EBSD patterns has been shown to be 95% accurate at a CI of 0.1 [36]. This threshold is then used to create a “mask” that flags individual voxels as containing good or bad (noisy) data, so the mask represents a subset of the total volume characterized during data collection. The mask for the sample of 304L can be seen in Fig. 3. Grains were segmented using a misorientation tolerance as described in “ Misorientation Tolerance Parameterization for Feature Identification,” and grains containing less than 64 voxels were considered too small to be resolved and were removed. This corresponds to 4 × 4 × 4 points across a feature and an equivalent diameter of 14.9 μm, roughly five times smaller than the average grain size determined from 2D analysis done prior to 3D characterization. Gaps remaining from these removed features were filled via iterative dilation of the surrounding grains. Indexed EBSD data was uploaded to BisQue for analysis after data collection.
EBM Inconel 718
Electron beam melting (EBM) is a powder-bed additive technique which utilizes an electron beam as a heat source. A fine layer of powder on the order of 50–100 μm is spread over a build plate, and the electron beam selectively melts the powder. The build plate is then lowered and the process is repeated to create complex geometries in 3D. Unlike laser melting techniques, electron beam melting requires vacuum processing environment, and the electron beam heat source can also be used to achieve preheat temperatures in excess of 1000 ∘C. A sample of EBM Inconel 718 was fabricated with 50-μm-thick build layers and was characterized in the TriBeam system with 1.5-μm-thick slices as described previously [37], and EBSD data was collected to characterize grain texture and morphology. Roughly 25 GB of data was collected from a total of 204 slices over the course of three days, resulting in a volume of 952.5 × 501.0 × 306.0 μm. Raw EBSD patterns were not collected, but the Hough transforms of the patterns were saved for post-processing. Glancing-angle FIB was not required to resolve EBSD patterns, which were collected with an in-plane resolution of 1.5 μm to create square voxels. As with the 304L sample, a minimum CI of 0.1 was chosen for the initial mask. Grains were segmented using a misorientation tolerance of 2∘ using the same segmentation algorithm as explained in “Misorientation Tolerance Parameterization for Feature Identification.” A minimum size requirement of 125 voxels was applied to delete grains that were too small to be properly characterized. This corresponds to 5 × 5 × 5 points across a feature and an equivalent diameter of 9.3 μm, roughly five times smaller than the average grain size determined from 2D analysis done prior to 3D characterization. Gaps remaining from these removed features were filled via iterative dilation of the surrounding grains. Indexed EBSD data was uploaded to BisQue for analysis after data collection.
Results
Misorientation Tolerance Parameterization for Feature Identification
In order to analyze 3D tomography data, individual features, or grains, must be segmented from the reconstructed volume. When working with 3D EBSD data, orientation information exists at every voxel in the volume. Straightforward segmentation algorithms such as connected component analysis (CCA) are used to identify grains from the 3D data using a misorientation tolerance between adjacent voxels. The misorientation tolerance is one of the most critical parameters for the segmentation and analysis of EBSD data. A mask is defined that distinguishes good data from noise in the dataset by using a threshold on the electron backscatter pattern confidence index (CI) [36]. To perform grain segmentation, a seed is selected from voxel 0 within the mask volume and its orientation is compared to each of its nearest neighbors that are still within the mask (up to six neighbors in a rectilinear voxelized grid). If the misorientation between two adjacent voxels is below the user-defined misorientation tolerance, then the voxel is added to the current grain. This process is repeated until no additional neighboring voxels can be added to the grain. A new seed is then selected at the smallest voxel index not previously grouped and the process continues until all voxels in the mask have been assigned to some grain ID.
It is essential that an optimal misorientation tolerance value is used during dataset reconstruction due to the impact of the misorientation tolerance on the definition of grain structure. Two-dimensional analysis of EBSD data typically uses a misorientation tolerance of 5∘, which is generally sufficient for well-annealed or recrystallized microstructures of various morphologies [38]. However, in deformed or highly textured materials, as are commonly observed in as-built additive structures [39, 40], such large misorientation tolerances can inadvertently merge grains that should otherwise be segmented separately. The additional voxel connectivity provided by 3D data further exacerbates this problem, and as there is generally not an accepted misorientation tolerance that applies to all 3D microstructures, these values must be chosen intuitively and the resulting segmentation analyzed for efficacy. This is a time-consuming, laborious process, and the ultimate selection of a user-defined parameter is not necessarily verifiable as the optimal choice. The use of parameterization, enabled by the BisQue framework, allows for the generation of many 3D volumes to better assess these and other important reconstruction parameters.
Keeping all other pipeline parameters constant (minimum number of two neighbors, minimum allowed defect size of 125 voxels), the misorientation tolerance was tested over a wide range of values, from 0.1 to 10∘ of misorientation in 0.1∘ increments, generating a total of one-hundred 3D reconstructions of the steel sample described in “LENS-Processed 304L Stainless Steel.” This represents some seventy gigabytes of generated data. To assess the quality of the reconstruction, two high-level metrics were used, the fraction of “good” data, and the total number of grains found during the reconstruction. “Good” data is defined by the confidence index of the EBSD data, and therefore contains a constant number of voxels that does not change between reconstructions. During the reconstruction process, various cleanup steps are employed which retain varying amounts of this initial group of voxels. DREAM.3D filters such as “Minimum Size,” “Minimum Number of Neighbors,” and “Fill Bad Data” can affect whether individual voxels are retained or released from segmented features. At the end of a reconstruction, post-processing scripts are used to count the number of voxels belonging to segmented features, which will always be a subset of the initial group of “good” voxels.
As can be seen in Fig. 5a, these two metrics tend to have opposite trends with increasing misorientation tolerance. As larger misorientation tolerances are used to segment the data, not only are more grains merged together, decreasing the overall number of grains found, but noisier data on the sample surfaces can also be connected to existing grains during segmentation. This trend is true except for the lowest misorientation tolerances, below 0.9∘. At these lower misorientation tolerances, the minimum grain size requirement of 64 voxels eliminates many of the grains found during the segmentation step, and for tolerances below 0.4∘, no continuous grains larger than 64 voxels are found, so grain-level metrics do not exist in these reconstructions.
An optimized reconstruction will maintain a high fraction of the masked voxels with high confidence index, while also reducing unnecessary merging of grains. The merging of grains when a large misorientation tolerance is selected can be seen in Fig. 4. The feature in Fig. 4a is identified as a single grain when using a misorientation tolerance of 5.0∘, but becomes separated into three separate grains when using a misorientation tolerance of 2.0∘ (Fig. 4a). The further separation of this feature results in grains with more reasonable grain morphologies that reflect the additive process. The best misorientation tolerance value is the lowest value at which both the change in fraction of masked data included and the change in the number of grains became constant, or when the derivatives of these quantities with respect to misorientation tolerance begin to converge, similar to the Elbow method used for determining the optimal number of clusters in a dataset. The derivatives are approximated via the central difference method, shown in Fig. 5b and c. A best balance between these metrics occurs at 2.0∘, making this an equitable choice of misorientation tolerance for segmentation of this dataset to avoid excess consolidation of separable grains. This results in a dataset with 899 grains, wherein 98.4% of the masked data is included in the reconstruction.
a A single feature identified from the bulk using a misorientation tolerance of 5.0∘ for segmentation via CCA. Grain orientation is shown in IPF coloring with the build direction taken as the reference direction. b The same feature is split into three separate grains when a misorientation tolerance of 2.0∘ is used for segmentation
a The fraction of the voxels in the confidence index mask included in the segmentation and the total number of grains, both as a function of the defined grain segmentation misorientation tolerance. The rate of change per degree of misorientation tolerance for b the fraction of included mask voxels and c the total number of grains. The optimal misorientation tolerance occurs at 2.0∘, where the derivatives start to converge
Subvolume Creation and Analysis of Large 3D Volumes
Beyond investigation of individual pipeline parameters, the BisQue framework also enables coupled parameterization, wherein sets of variables are linked to each other. This allows for automated generation of random samples of equal-sized subvolumes of 3D datasets, among other applications. The characterization of subvolumes in 3D datasets has been previously applied to define microstructural and property volume elements (MVEs and PVEs) for various material properties, to inform component design [41, 42].
Morphological Complexity of As-printed Additive Structures
Due to the complex thermal gradients and remelting phenomena inherent to additive manufacturing methods, additive microstructures are highly variable and can be morphologically complex [43,44,45,46]. As previously noted, these heterogeneous microstructures can easily be misinterpreted in standard analyses of 2D cross-sections [47]. Columnar grains near the center of melt pool, when cut perpendicularly, can appear as fine equiaxed grains, which may mimic traditional weld microstructures and can lead to misinterpretation of data. Misinterpretation is also highly likely for larger grains in additive processes. Either as a result of multiple remelting and solidification cycles, or from a single solidification event with complex thermal gradients, individual grains are commonly observed to possess large misorientation gradients. One such large grain was identified in the reconstruction of the 304L stainless steel sample described in “Misorientation Tolerance Parameterization for Feature Identification.” Even while using the previously determined misorientation tolerance of 2∘ for grain segmentation, large changes in orientation are observed, in excess of 12∘ from the average orientation of the grain. Changes in orientation are evident via differences in inverse pole figure (IPF) coloring of the grain shown in Fig. 6.
a A large grain in LENS-processed additive manufactured 304L stainless steel with misorientation gradients. Three orthogonal slicing directions illustrating how the microstructure may be misinterpreted by traditional 2D metallurgical sample preparation and sectioning include b X-normal slices, c Y-normal slices, and d Z-normal slices
The limitations of 2D characterization of additive structures such as those shown in Fig. 6 are demonstrated by examining 2D cross-sections from the 3D data volume. Using the coupled parameterization ability of BisQue and the “Crop Geometry” filter in DREAM.3D, individual subvolumes, each 1 voxel thick (to approximate 2D cross-sections), were extracted from a 3D grain along the three principal sample directions (Fig. 6b–d). Figure 6d shows Z-normal slices, which are those taken with a plane normal along the build direction of the sample, which is also parallel to the TriBeam sectioning direction. X-normal (Fig. 6b) and Y-normal (Fig. 6c) slices are those containing a plane normal along either of the in-plane axes as defined by the TriBeam sectioning direction and the sample geometry. These slices were then segmented using the same misorientation tolerance used on the full volume, 2∘, to determine how the lack of 3D data would affect the characterization of such a grain. As this analysis was only performed on voxels associated with a large feature from the 3D volume, features resulting from this additional segmentation are subfeatures of the initial 3D grain. An example of a Z-normal slice characterization is shown in Fig. 7. Since small features comprised of fewer than 64 voxels were already removed from the 3D volume, a modest minimum size requirement of four pixels was applied to these slices, effectively removing what would normally be treated as noisy pixels in typical 2D cross-sections. The 3D nature of this large grain can result in segmented subfeatures that appear isolated from the rest of the original feature in a single slice, but in fact connect out of plane. A summary of the results from this analysis is shown in Fig. 8, where the number of subfeatures found in a single cross-section, the fraction of slice data contained within the largest subfeature, and the stereologically corrected equivalent grain diameter calculated from the largest subfeature are shown. A slice that accurately represents the 3D data will lie in the upper left portion of the graph and have a larger predicted equivalent diameter, closer to that determined from the 3D dataset.
Summary of 2D analysis of a large 3D grain. The number of subfeatures found in a single 2D cross-section, as well as the predicted grain size from the largest slice subfeature, is highly dependent on the location and orientation of the cross-section. A 2D slice that more accurately represents the true 3D structure will lie in the upper left portion of the graph, having a larger predicted equivalent diameter
Of the 467 slices generated, only 32 (6.85%) of the slices retained a single feature, and 76.7% of slices were found to contain five or more subfeatures, despite all of the data having been obtained from a single 3D grain. Due to the connectivity information that is missing in 2D, the gradual changes in orientation that exist in the material are interpreted as higher-angle grain boundaries (Fig. 7), resulting in their separation during segmentation via CCA. Similarly, attempts to characterize effective grain size of additive materials are inconsistent when working with 2D data. For example, the equivalent diameter of the large grain was found to be 320.0 μm as determined from the 3D reconstruction. Taking the largest subfeature found in a particular slice, the equivalent diameter of the grain can also be calculated from the 2D cross-sections. The 2D cross-sections under-predict the grain size as measured from the full volume of the dataset, with a median equivalent diameter of 189 μm. There is also a clear dependence on the location and orientation of the slice in the calculation of this value, with X- and Y-normal slices yielding median equivalent diameters of 156 and 155 μm, respectively, whereas the Z-normal slices have a median equivalent diameter of 267 μm. X- and Y-normal slices also show similar variability in the number of subfeatures found during 2D analysis, as well as the relative fraction of data contained in the largest subfeature. The distinctive trend of Z-normal slices, namely larger predicted equivalent diameters and proportionally larger subfeatures, indicates that these slices contain a larger number of small subfeatures, which can be seen in Fig. 7. These generated cross-sections reveal the inadequacy of 2D analysis to accurately assess grain size and shape, and emphasize the importance of 3D characterization in additively manufactured materials. The directional dependence of calculated properties also highlights the anisotropic properties that exist as a result of the microstructure in additively manufactured materials.
Anisotropic Defect Generation in Additive Processes
Characterization of a sample of EBM Inconel 718, as described in “EBM Inconel 718,” revealed a large, lack of fusion defect. The lack of fusion event resulted in the formation of large clusters of equiaxed grains above the defect, creating microstructural inhomogeneity in the otherwise columnar microstructure that cannot be easily mitigated via post-processing heat treatments, even if the porosity is closed from hot isostatic pressing (HIP) [37]. The clusters of equiaxed grains also lacked the strong texture present in the columnar grains that comprise the bulk of the volume. Quantification of clustering behavior of equiaxed grains was performed on a slice-per-slice basis through the volume, along the build direction. What was previously a time-intensive process to characterize a single lack of fusion defect can now be easily performed on the entire collected volume using a coupled-slicing procedure in BisQue as described in “Morphological Complexity of As-printed Additive Structures.” The clustering of equiaxed grains was quantified by comparing the area fraction of equiaxed grains in a single slice to the average volume fraction of equiaxed grains in the entire volume (Fig. 9). A basic relationship of stereology states that in regular, homogeneous structures, the volume fraction of a given phase or feature type in 3D is equivalent to measured areas in cross-section [48]. Thus, discrepancies between the measured area fraction of equiaxed grains in individual 2D cross-section and the volume fraction of these grains from 3D characterization arise due to heterogeneity in the microstructure.
The percent increase in equiaxed grains from dataset average in an AM Inconel 718 alloy as a function of dataset slice location along the build direction. Several secondary electron micrographs from the volume exhibit porosity, and in each case equiaxed grains lie above these defects along the build direction. These equiaxed grains can be seen via a loss of the <001> texture in the region above the defects in the EBSD data. The location of the secondary electron images and EBSD data are highlighted
The extent of these deviations from a fully heterogeneous microstructure are plotted in Fig. 9. Local maxima in the curve correspond to local clusters of equiaxed grains. The width of the associated peaks are consistently smaller than the build layer thickness of 50 μm, indicating that all of these clusters appear on individual build layers. Analysis of the entire volume shows that several clusters of equiaxed grains exist throughout the volume beyond the large cluster previously identified. In every case, clusters of equiaxed grains appear in the build layer above the lack of fusion defect. The change in thermal gradients arising from the lack of fusion result in formation of equiaxed grains that interrupt the otherwise columnar grain morphology observed in the bulk of this sample [37]. As successive build layers are deposited above the lack of fusion defects, columnar growth can resume, but equiaxed grains persist above the porosity. Secondary electron images taken during TriBeam tomography confirm that these clusters are spatially correlated to planar porosity defects occurring during part fabrication (Fig. 9). Many of these planar voids are < 5μm thick, below the detectable limit of lab-scale X-ray computed tomography and microtomography methods [49], so would not be readily identified with nondestructive evaluation techniques, despite their impact on changing the local microstructure morphology and texture. Quantitative analysis of the EBSD data in BisQue reveals that these smaller lack of fusion defects are much more prevalent throughout the sample, beyond the more easily identifiable, large-scale defect previously characterized.
Discussion
The ability of the BisQue platform to integrate materials science tools such as DREAM.3D can significantly advance the emerging 3D and 4D characterization communities. As the routines used to segment, reconstruct, clean, and analyze higher-dimensional data have not yet reached technical maturity, it is essential to develop tools that allow for simple and scalable exploration of parameter space offered by these data. This approach is especially important for certain classes of materials that are not easily analyzed through conventional segmentation algorithms. Additively manufactured microstructures in particular result from compound melting and solidification cycles, as well as associated solid-state transformations in the thermal histories of these parts. The complex grain morphologies and long-range orientation gradients present in additive microstructures cannot be accurately assessed using conventional techniques. Unlike reconstruction and analysis of 3D data based on trial-and-error efforts, adopting necessarily simplified approaches that cannot guarantee fidelity of an individual analysis, BisQue enables dataset-specific reconstruction parameters to be rigorously defined and studied. To that end, parallelization methods for assessing reconstruction fidelity offer a path forward for standardizing 3D materials characterization in a materials agnostic form; there may be no “one-size-fits-all” misorientation tolerance for segmentation of 3D EBSD data, but there could be a “one-size-fits-all” approach to determining the optimal value, streamlining the process of 3D materials analysis and more accurately representing these rich datasets. Such parameter studies could be extended to entire 3D reconstruction pipelines in DREAM.3D, rather than focusing on individual reconstruction parameters. Combined with other services in BisQue, such as a volume and table viewer compatible with common 3D data formats, much of the 3D reconstruction and analysis process is already possible completely through a web browser. Development of 3D analysis tools such as DREAM.3D for BisQue could take the entire process off of individual workstations, simplifying the process of 3D data analysis. Capabilities for raw data storage, reconstruction, and analysis of 3D microstructures discussed here are also useful because they can be combined with tools for predicting microstructure-sensitive properties. Such tools which may include analytic bounds (e.g., Voigt/Reuss) or emerging data-driven models for microstructure-property relationships [50] can also be incorporated into BisQue, which is discussed in detail elsewhere [51].
The scalable infrastructure of BisQue offers numerous advantages to the materials science community. Scale-up and replication of BisQue servers is achievable using well-known methods such as reverse proxies and replicated servers. Leveraging high-performance computing capabilities provides the potential to process very large datasets in a distributed manner. Accessed via a web-browser, BisQue enables efficient and straightforward use of complex analysis algorithms with minimal software and hardware requirements on the user end. Underlying all of this is continual updating and tracking of metadata, whose structure and context can be formulated to cater to specific research community needs, maintaining high flexibility of the infrastructure. A core component of metadata is the retention of data provenance at every stage of the process. When a resource is created or modified, it is marked by the system, so a user can track the full history of a dataset from its creation, even when shared among several users. This fosters collaboration and allows multiple researchers to analyze and edit the same dataset without fear of modifying the original raw data. Due to the highly specialized equipment and facilities required to generate 3D materials data, 3D materials science is necessarily collaborative. The ability to share and distribute large datasets around the globe, as well as the associated analyses, is critical to the growth of the 3D materials community at large.
A distributed computing infrastructure can also benefit the need to certify additively manufactured parts and characterize the build process, which requires powerful analytic methods. During operation, additive manufacturing machines record a myriad of variables simultaneously, ranging from the power and location of the heat source to chamber pressure and moisture content, all of which are correlated to observed microstructures and defect generation [52]. The use of specialized in situ sensors, such as near-IR (infrared) imaging for defect detection, thermographic measurement of temperature profiles, or high-speed imaging to investigate fluid flow and powder dynamics are essential to advancing our understanding of additive manufacturing techniques [53,54,55,56]. At the same time, the emergence of these techniques increases the complexity of data associated with individual additively manufactured parts. Not all sensors record data at equivalent rates, and many do not use regular sampling intervals. Data visualization tools such as FALCON have been specifically designed to mitigate these issues and enable machine users to determine processing-property linkages [57]. New high-throughput ex situ characterization of additively manufactured parts has also demonstrated the sensitivity of part performance to build parameters and machine-to-machine variability [58, 59], underlying the need for a more comprehensive understanding of the build process. Of particular concern to additive material qualification are the so-called rare events at extrema of property distributions not easily seen without such high-throughput testing, severely limiting confidence in the application of this technology to critical components [60]. Incorporation of such tools, in combination with advanced statistical approaches and emerging machine-learning algorithms from other communities, into a collaborative environment such as BisQue can help advance these algorithms and grow user communities. Coupling of BisQue to additive machines could enable real-time experimental analysis and provide feedback to machine operators. Distributing analysis and collection of build data and part properties allows users to leverage advanced sensor capabilities, enabling additive manufacturing to become a self-correcting, adaptive process that can deliver “born qualified” parts [61].
As tools for BisQue develop, there is enormous opportunity to support growing research trends. Integration with GitHub repositories would enable version management of custom analytic tools, providing archival management of developing codebases. The use of dockerized environments for running such software in BisQue ensures that datasets analyzed using out-of-date code will still function properly, and old analyses will be preserved. Annotation on datasets can be utilized as a new approach for performing collaborative analysis and marking data for automated analysis. New analysis tools and custom scripts for common packages such as ImageJ and Jupyter notebooks can constantly be updated and linked via BisQue’s data provenance construct. Automated analysis for specific data structures could be implemented, akin to facial recognition processes performed by Google’s Vision API on images as they are uploaded to the Cloud. BisQue could also serve as a repository for published datasets, similar to the databases such as Mendeley Data and Journals such as Data in Brief, Scientific Data, and Data which publish scientific datasets, providing a Digital Object Identifier (DOI) and enabling their use by other researchers. Beyond sharing the data, however, BisQue can fully track the usage and analysis of the data all the way back to its source. Particularly for 3D datasets, which contain a wealth of information, BisQue can enable multiple research groups with diverse specialties to work on the same data, providing new insights and fostering new connections among researchers.
Conclusion
The 3D microstructure reconstruction and analysis software DREAM.3D was integrated as a module into the BisQue cloud-based platform allowing for the following:
-
Archival preservation of data provenance
-
High-performance cluster computing integration (parallelized DREAM.3D module execution)
-
Parameterization of DREAM.3D pipelines for reconstruction optimization
-
Algorithms for the 2D and 3D volume element sampling from large 3D datasets
The DREAM.3D pipeline parameterization combined with parallelized module executions has allowed for the exploration and optimization of dataset reconstructions. For example, new insights into the choice of grain segmentation misorientation tolerances for additively manufactured materials were discovered. Using algorithms for the virtual 2D and 3D volume element sampling from large 3D datasets, traditional 2D metallographic sectioning was found to yield grain size measurements that were roughly half the size of the equivalent 3D measurements in heterogeneously distributed microstructures such as those found in AM materials. These measurements were performed with a massively parallelized array of slicing/cropping pipelines to simulate the 2D vs 3D statistical grain measurements.
References
US Department of Energy (2014) Public access plan. Technical report, US Department of Energy, Washington
National Science Foundation (2015) Today’s data, tomorrow’s discoveries. Technical report, National Science Foundation
Ward C National Materials Data Initiatives. In 2015: UCSB workshop on collection and analysis of big data in 3D materials science, Santa Barbara
Gewin V (2016) An open mind on open data, vol 529
The Minerals, Metals & Materials Society (TMS) (2017) Building a materials data infrastructure: opening new pathways to discovery and innovation in science and engineering. Technical report, TMS, Pittsburgh PA
Jain A, Ping Ong S, Hautier G, Chen W, Richards WD, Dacek S, Cholia S, Gunter D, Skinner D, Ceder G, Persson KA (2013) The materials project: a materials genome approach to accelerating materials innovation. APL Mater 1(1):011002
Saal JE, Kirklin S, Aykol M, Meredig B, Wolverton C (2013) Materials design and discovery with high-throughput density functional theory: the open quantum materials database (oqmd). JOM 65(11):1501–1509
Groeber MA, Jackson MA (2014) Dream.3d: a digital representation environment for the analysis of microstructure in 3d. Integr Mater Manuf Innov 3(1):5
Adams BL, Wright SI, Kunze K (1993) Orientation imaging: the emergence of a new microscopy. Metall Trans A 24(4):819–831
Friel J, Wright S, Sitzman S (2011) ASTM grain size by EBSD - a new standard. Microsc Microanal 17(S2):838–839
ASTM E2627-13 (2013) Standard practice for determining average grain size using electron backscatter diffraction (EBSD) in fully recrystallized polycrystalline materials. ASTM International, West Conshohocken, PA
Humphreys FJ (2004) Characterisation of fine-scale microstructures by electron backscatter diffraction (EBSD). Scr Mater 51(8):771–776
Mingard KP, Roebuck B, Bennett EG, Thomas M, Wynne BP, Palmiere EJ (2007) Grain size measurement by EBSD in complex hot deformed metal alloy microstructures. J Microsc 227(3):298–308
Geiss RH, Read DT (2007) Need for standardization of EBSD measurements for microstructural characterization of thin film structures. In: AIP conference proceedings, vol 931, pp 168–172. AIP
Mingard KP, Roebuck B, Bennett EG, Gee MG, Nordenstrom H, Sweetman G, Chan P (2009) Comparison of EBSD and conventional methods of grain size measurement of hardmetals. Int J Refract Met Hard Mater 27(2):213–223
Sun L, Thomas MJ, Wynne BP, Palmiere EJ, Mingard K P, Roebuck B (2010) Mapping microstructure inhomogeneity using electron backscatter diffraction in 316L stainless steel subjected to hot plane strain compression tests. Mater Sci Technol 26(12):1477–1486
Latypov MI, Ku̇hbach M, Beyerlein IJ, Stinville J-C, Toth LS, Pollock TM, Kalidindi SR (2018) Application of chord length distributions and principal component analysis for quantification and representation of diverse polycrystalline microstructures. Mater Charact 145:671–685
Bourdin F, Stinville JC, Echlin MP, Callahan PG, Lenthe WC, Torbet CJ, Texier D, Bridier F, Cormier J, Villechaise P et al (2018) Measurements of plastic localization by heaviside-digital image correlation. Acta Mater 157:307–325
Wright SI, Nowell MM, Field DP (2011) A review of strain analysis using electron backscatter diffraction. Microsc Microanal 17(03):316–329
Britton TB, Holton I, Meaden G, Dingley DJ (2013) High angular resolution electron backscatter diffraction: measurement of strain in functional and structural materials. Microscopy and Analysis, (May): 1–5
Stinville JC, Echlin MP, Texier D, Bridier F, Bocher P, Pollock TM (2016) Sub-grain scale digital image correlation by electron microscopy for polycrystalline materials during elastic and plastic deformation. Exp Mech 56(2):197–216
Wilkinson AJ, Britton TB, Jiang J, Karamched PS (2014) A review of advances and challenges in EBSD strain mapping. IOP Conference Series: Materials Science and Engineering 55(1):012020
Vaudin MD, Osborn WA, Friedman LH, Gorham JM, Vartanian V, Cook RF (2015) Designing a standard for strain mapping: HR-EBSD analysis of SiGe thin film structures on Si. Ultramicroscopy 148:94–104
Ludwig W, Schmidt S, Lauridsen EM, Poulsen HF (2008) X-ray diffraction contrast tomography: a novel technique for three-dimensional grain mapping of polycrystals. I. Direct beam case. J Appl Crystallogr 41(2):302–309
Reischig P, King A, Nervo L, Viganó N, Guilhem Y, Palenstijn WJ, Batenburg KJ, Preuss M, Ludwig W (2013) Advances in X-ray diffraction contrast tomography: flexibility in the setup geometry and application to multiphase materials. J Appl Crystallogr 46(2):297–311
Groeber MA, Haley BK, Uchic MD, Dimiduk DM, Ghosh S (2006) 3D reconstruction and characterization of polycrystalline microstructures using a FIB–SEM system. Mater Charact 57(4-5):259–273
Bobrowski P, Pėdzich Z, Faryna M (2015) Three-dimensional microstructural characterization of porous cubic zirconia. Micron 78:73–78
Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS (2009) Bisque: a platform for bioimage analysis and management. Bioinformatics 26(4):544–552
Kvilekval K, Fedorov D, Gaur U, Goff S, Merchant N, Manjunath BS, Singh AK (2012) Bisque: advances in bioimage databases. IEEE Data Eng Bull 35(3):56–64
Echlin MP, Straw M, Randolph S, Filevich J, Pollock TM (2015) The tribeam system: femtosecond laser ablation in situ sem. Mater Charact 100(0):1–12
The HDF Group Hierarchical data format, version 5, 1997-NNNN. http://www.hdfgroup.org/HDF5/
Ahrens J, Geveci B, Law C (2005) Paraview, an end-user tool for large data visualization. In: Visualization handbook. Elsevier
Echlin MP, Mottura A, Torbet CJ, Pollock TM (2012) A new TriBeam system for three-dimensional multimodal analysis. Rev Sci Instrum 83(2):023701
Echlin MP, Titus MS, Straw M, Gumbsch P, Pollock TM (2017) Materials response to glancing incidence femtosecond laser ablation. Acta Mater 124:37–46
Brockmann R, Candel-Ruiz A, Kaufmann S, Müllerschön O (2015) Strategies for high deposition rate additive manufacturing by laser metal deposition. In: International congress on applications of lasers & electro-optics, vol 680, pp 680–683. Laser Institute of America
Field DP (1997) Recent advances in the application of orientation imaging. Ultramicroscopy 67(1-4):1–9
Polonsky AT, Echlin MLP, Lenthe WC, Dehoff RR, Kirka MM, Pollock TM (2018) Defects and 3D structural inhomogeneity in electron beam additively manufactured Inconel 718. Mater Charact 143:171–181
Wright SI (2010) A parametric study of electron backscatter diffraction based grain size measurements. Pract Metallogr 47(1):16–33
Dinda GP, Dasgupta AK, Mazumder J (2012) Texture control during laser deposition of nickel-based superalloy. Scr Mater 67(5):503–506
Dehoff RR, Kirka MM, List FA, Unocic KA, Sames WJ (2015) Crystallographic texture engineering through novel melt strategies via electron beam melting: Inconel 718. Mater Sci Technol 31(8):939–944
Echlin MLP, Lenthe WC, Pollock TM (2014) Three-dimensional sampling of material structure for property modeling and design. Integr Mater Manuf Innov 3(1):21
Lenthe WC, Stinville J-C, Echlin MLP, Pollock TM (2016) Statistical assessment of fatigue-initiating microstructural features in a polycrystalline disk alloy. In: Superalloys 2016: Proceedings of the 13th international symposium of superalloys, pp 569–577. Wiley Online Library
Al-Bermani SS, Blackmore ML, Zhang W, Todd I (2010) The origin of microstructural diversity, texture, and mechanical properties in electron beam melted Ti-6Al-4V. Metall Mater Trans A 41(13):3422–3434
Carter LN, Martin C, Withers PJ, Attallah MM (2014) The influence of the laser scan strategy on grain structure and cracking behaviour in SLM powder-bed fabricated nickel superalloy. J Alloys Compd 615:338–347
Wang YM, Voisin T, McKeown JT, Ye J, Calta NP, Li Z, Zeng Z, Zhang Y, Chen W, Roehling TT, Ott RT, Santala MK, Depond PJ, Matthews MJ, Hamza AV, Zhu T (2017) Additively manufactured hierarchical stainless steels with high strength and ductility. Nat Mater 17(1):63–71
Rodgers TM, Madison JD, Tikare V (2017) Simulation of metal additive manufacturing microstructures using kinetic Monte Carlo. Commun Math Sci 135(3):78–89
Wei HL, Elmer JW, DebRoy T (2017) Three-dimensional modeling of grain structure evolution during welding of an aluminum alloy. Acta Mater 126:413–425
Kurzydłowski JK, Ralph B (1995) The quantitative description of the microstructure of materials. CRC Press
Cunningham R, Narra SP, Ozturk T, Beuth J, Rollett AD (2016) Evaluating the effect of processing parameters on porosity in electron beam melted Ti-6Al-4V via synchrotron x-ray microtomography. JOM 68(3):765–771
Latypov MI, Toth LS, Kalidindi SR (2019) Materials knowledge system for nonlinear composites. Comput Methods Appl Mech Eng 346:180–196
Latypov MI, Khan A, Lang CA, Kvilekval K, Polonsky AT, Echlin MP, Beyerlein IJ, Manjunath BS, Pollock TM (2019) BisQue for 3D materials science in the cloud: microstructure–property linkages. https://doi.org/10.1007/s40192-019-00128-5
Groeber MA, Schwalbach E, Donegan S, Chaput K, Butler T, Miller J (2017) Application of characterization, modelling, and analytics towards understanding process-structure linkages in metallic 3D printing. IOP Conference Series: Materials Science and Engineering 219(1):012002
Dinwiddie RB, Dehoff RR, Lloyd PD, Lowe LE, Ulrich JB (2013) Thermographic in-situ process monitoring of the electron-beam melting technology used in additive manufacturing. Number May 2013, page 87050K
Lane B, Moylan S, Whitenton EP, Ma L (2016) Thermographic measurements of the commercial laser powder bed fusion process at NIST. Rapid Prototyp J 22(5):778–787
Matthews MJ, Guss G, Khairallah SA, Rubenchik AM, Depond PJ, King WE (2016) Denudation of metal powder layers in laser powder bed fusion processes. Acta Mater 114:33–42
Niyanth Niyanth S, Baba JS, Jordan BH, Dinwiddie RB, Dehoff Ryan R (2018) Understanding part to part variability during directed energy deposition processes using in-situ and ex-situ process characterization. Technical report
Steed CA, Halsey W, Dehoff R, Yoder SL, Paquit V, Powers S (2017) Falcon: Visual analysis of large, irregularly sampled, and multivariate time series data in additive manufacturing. Comput Graph 63:50–64
Salzbrenner BC, Rodelas JM, Madison JD, Jared BH, Swiler LP, Shen Y-L, Boyce BL (2017) High-throughput stochastic tensile performance of additively manufactured stainless steel , vol 241
Madison JD, Underwood OD, Swiler LP, Boyce BL, Jared BH, Rodelas JM, Salzbrenner BC (2018) Corroborating tomographic defect metrics with mechanical response in an additively manufactured precipitation-hardened stainless steel. In: AIP conference proceedings, vol 1949, pp 020009
Boyce BL, Salzbrenner BC, Rodelas JM, Swiler LP, Madison JD, Jared BH, Shen Y-L (2017) Extreme-value statistics reveal rare failure-critical defects in additive manufacturing. Adv Eng Mater 19(8):1700102
Roach AR, Abdeljawad F, Argibay N, Allen K, Balch D, Beghini L, Bishop J, Boyce B, Brown J, Burchard R, Chandross M, Cook A, Dressler A, Forrest E, Ford K, Ivanoff T, Jared B, Kammler D, Koepke J, Kustas A, Lavin J, Leathe N, Lester B, Madison J, Mani S, Martinez MJ, Moser D, Murphy R, Rodgers T, Seidl T, Shaklee-brown H, Stanford J, Stender M, Sugar J, Swiler LP, Taylor S, Trembacki B, Van Bloemen Waanders B, Whetton S, Wildey T, Wilson M (2018) Born qualified grand challenge LDRD final report. Technical Report September, Sandia National Laboratories
Acknowledgments
The authors would like to acknowledge the support of NSF EAGER Grant No. 1650972 and NSF SI2-SSI Grant No. 1664172. Andrew T. Polonsky and Tresa M. Pollock also acknowledge Ryan R. Dehoff and Michael M. Kirka for providing additive material and Oak Ridge National Laboratory for support under Award No. 400156470 and acknowledge George T. Gray III and Veronica Livescu of Los Alamos National Laboratory for providing additive material. The authors acknowledge Mike Jackson from BlueQuartz software for DREAM.3D discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Polonsky, A.T., Lang, C.A., Kvilekval, K.G. et al. Three-dimensional Analysis and Reconstruction of Additively Manufactured Materials in the Cloud-Based BisQue Infrastructure. Integr Mater Manuf Innov 8, 37–51 (2019). https://doi.org/10.1007/s40192-019-00126-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-019-00126-7