Abstract
Machining is a severe plastic deformation process, wherein the workpiece material is subjected to high deformation rates and temperatures. During metal machining, the dynamic recrystallization mechanism causes grain refinement into the sub-micron range. In this study, we investigate the microstructure evolution of oxygen-free high conductivity copper (OFHC Cu) subject to a machining process where the cutting speed and rake angle are controlled to manipulate the process strain, strain rate, and temperatures. Microstructures of the deformed chips are quantified using orientation imaging microscopy and novel statistical descriptors that capture the morphology and local lattice misorientations generated during the several mechanistic stages of the dynamic recrystallization process. Mechanical properties of the resulting chips are quantified using spherical nanoindentation protocols. A multiple output Gaussian process regression model is used to simultaneously model the structure-property evolution, which differs from more common approaches that establish such relationships sequentially. This modeling strategy is particularly attractive since it can flexibly provide both structure and property uncertainty estimates. In addition, the statistical modeling framework allows for the inclusion of multi-fidelity data. The statistical metrics utilized serve as efficient microstructure descriptors, which retain the physics of the observed structures without having to introduce ad hoc microstructure feature definitions.
Similar content being viewed by others
Introduction
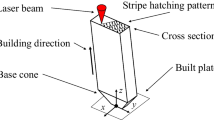
Machining is a high-rate severe plastic deformation (SPD) manufacturing process. The process can be described using the idealized model shown in Fig. 2a. The imposed thermomechanical loading is fairly extreme with imposed strains as large as \(\gamma \sim 10\), deformation rates up to 105s− 1, and cutting temperatures as high as 0.6𝜃 (homologous temperature) [1]. These imposed deformation conditions result in microstructure refinement in both the deformed chip and the component surface [2,3,4,5,6, 6,7,8]. The corresponding mechanical properties of both the chip and the workpiece surface are naturally sensitive to the produced structures [3, 8,9,10]. Therefore, identifying the process-structure-property (PSP) relationships that characterize machining is critical for establishing a synergistic framework where designers, materials scientists, and manufacturers can cooperate to engineer functional surfaces. PSP relationships are often expressed via mechanism maps as shown in Fig. 1. Furthermore, the SPD structures produced in machining bear a resemblance to structures produced in other SPD processes, such as equal channel angular extrusion [11], high pressure torsion [12], and dynamic processes where shear banding may occur [13,14,15]. Therefore, the merit in studying machining as a high-rate SPD process translates to other fields as well.
Machining process-structure-property map [7]
The predominant microstructure evolution mechanism in machining under ambient conditions is either continuous or discontinuous dynamic recrystallization (CDRX or DDRX) [3, 9]. CDRX is driven by the formation of dislocation cells that transform to low angle boundary (LAB) sub-grain structures, and finally relative sub-grain rotations generate high angle boundary (HAB)-refined grains [9, 16]. DDRX is more closely related to classic recrystallization where new grains nucleate and grow, often near existing grain boundaries [17]. Since the mechanism driving CDRX is driven by lattice rotations, the structure evolution can be quantified by considering measures of crystallographic misorientation [8, 18,19,20,21]. Mechanical constitutive property measurements are usually limited to hardness since the produced samples are small in scale (machined chips and workpiece surface) [3, 7,8,9,10].
Materials informatics (MI) is an emerging field within the materials community which, like cheminformatics and bioinformatics, seeks to employ statistics for addressing important domain science problems [22,23,24,25]. Materials research is conducted utilizing statistical approaches for establishing data-driven models, quantifying uncertainty, and the design and planning of experiments. MI addresses the fundamental challenge in materials research, identifying PSP relationships, by building mathematically rigorous models. The models, which may be data-driven or mixed data/physics models, may then be exploited for the design of functional materials. Recent works have established reduced-order structure-property (SP) models for single-phase polycrystalline systems [26, 27]. These authors utilized generalized spherical harmonics (GSH) to quantify bulk textures and used spatial statistics to quantify the spatial structure describing various simulated microstructure realizations. Another recent work utilized a deep adversarial learning model coupled with a Gaussian process (GP) Bayesian design criteria for computational materials design [28].
In this work, we study the evolution of pure copper subject to a high-rate SPD machining process. Microstructure is quantified using orientation imaging microscopy (OIM). A microstructure statistic which quantifies the local crystal spatial misorientation is derived. This is done by utilizing a GSH basis to describe the crystallographic orientation and a unique spatial autocorrelation function, which exploits the orthogonality of the GSH basis. Constitutive mechanical properties are quantified using spherical nanoindentation tests. Finally, a multiple output Gaussian process regression (MOGPR) model is developed, which captures the full PSP relationships as well as their associated uncertainties. The model is flexible and is well suited for handling multiple kinds of data, e.g., multi-fidelity modeling.
Experimental Methods
Oxygen-free high conductivity copper (OFHC Cu) bars were obtained from a supplier (McMaster Carr). The material was subjected to SPD via a machining process. Tube turning experiments were carried out to emulate the idealized two-dimensional orthogonal cutting experiment shown in Fig. 2a. High speed steel cutting tools with nominal rake angles α = 5∘,15∘,25∘,45∘ were used for all experiments. A constant feed (or undeformed chip thickness) (to) of 300μ m was prescribed for all tests. The prescribed geometry was chosen to impose large shear strains in the primary shear zone, which the machining theory predicts to be \(\gamma \sim 1-8\) [1]. Four cutting speeds V = 0.20,0.33,0.50,1.00m ⋅s− 1 were studied, which generate strain rates \(\sim 10^{3}- 10^{4} \text {s}^{-1}\). Higher cutting speeds correspondingly yield increases in chip temperatures as there is less time available for diffusion of heat away from the chip. From the measured cutting forces, the chip temperatures were estimated to reach \(\sim 165~^{\circ }\text {C}\) for the lowest rake angle (highest strain) and fastest cutting speeds utilized [1]. Generated chips fell into a quench tank filled with water to freeze the as-machined microstructure.
Collected chips were mounted in epoxy as shown in Fig. 2b. Small sample-to-sample deviations in the chip orientation within the casting will affect the perceived two-dimensional morphology of observed micrographs. Furthermore, OIM results will be affected due to uncertainty in the reference sample orientation. Therefore, special care was taken to mount samples such that the observed cross section correspond as closely as possible to the idealized two-dimensional orthogonal configuration. Grinding of the metallographic samples was performed to reach the “mid chip” (1mm) thickness which is far from free boundaries and therefore minimally affected by side flow transverse to the direction of chip flow. Samples were subsequently mechanically polished with up to 1μ m diamond suspension polish. Final surface preparation was performed via vibratory polishing in a Buehler VibroMet 2. A Tescan Mira XMH field emission scanning electron microscope (FE-SEM) was utilized to image the generated microstructures. A backscatter emissions (BSE) detector was utilized for all imaging as it was found to yield images with extremely good contrast (see, Fig. 3). A EDAX Hikari EBSD detector with TSL OIM analysis was utilized for orientation imaging (Fig. 1).
Nanoindentation experiments were performed on a Agilent G200 nanoindenter with an XP head and continuous stiffness monitoring (CMS). A 100μ m diamond indenter was used for all experiments. Spherical indentation stress-strain protocols were utilized to further process experimental data [29, 30]. The derived indentation stress-strain curves capture the mechanical response of the material deformed beneath the indenter. The corresponding contact radius for these experiments varied between 10 and 20 μ m. The microstructures considered vary greatly in their degree of refinement. Under some conditions, very fine structures (d < 1μ m) were generated suggesting that the obtained indentation responses are likely well homogenized. Coarser structures, however, suggest that the local material heterogeneity may introduce additional response variation. In our analysis, we will account for this by attempting to establish mean property quantities.
Microhardness measurements were performed using a Buehler series 1600 microhardness tester. A diamond tip Vickers indenter loaded to 500 g was used for all tests.
Methods
Microstructure Quantification
BSE and EBSD micrographs for two different process conditions are shown in Fig. 3. Images at larger values of the rake angle α (or smaller values of strain since γ ∝ α− 1) produced correspondingly coarser microstructures; therefore, larger fields of view were required at these settings. The field of view at each setting is illustrated in Fig. 4. The total number of raster steps in each image was maintained at 300 × 300 to avoid unnecessarily long scans.
In Fig. 3, it is clear from both the BSE and EBSD scans that the microstructures are morphologically different. In the α = 25∘ BSE image, however, it is difficult to discern which features are grain boundaries; the BSE image is sensitive to defect structures besides grain boundaries. An even clearer pattern is visible in Fig. 4, particularly at low rake angles of 5∘ and 15∘; with increasing cutting speed, it appears as if the structure becomes smeared. Statistically, it can be stated that crystal orientations are more spatially correlated at higher cutting speeds than at lower cutting speeds. Furthermore, this pattern is also present with increasing rake angle. Consider an experiment where a point is chosen randomly in the micrograph for 5∘,1.00 m ⋅s− 1, and we note the crystal orientation at the chosen pixel and at a location 5 μ m to its right. Subsequently, the same experiment is performed on the micrograph for (5∘,0.20 m ⋅s− 1). On average, over many repetitions, the two pixels from 5∘,1.00 m ⋅s− 1 would yield more “similar” orientations than in the micrograph for 5∘,0.20 m ⋅s− 1. It is this feature that we wish to quantify and exploit for assessing microstructural anisotropy.
Recent advances in the MI community have established statistically rigorous methods for quantifying stochastic material systems [24]. In this work we quantify microstructure via crystallographic orientation which can be quantified using the Bunge-Euler angles \(\boldsymbol {g}=\left (\phi _{1},{\Phi },\phi _{2}\right )\), which are continuously defined over the fundamental zone (FZ) [31]. The probability of finding orientation g at spatial location x is \(f_{x}\left (\boldsymbol {g}\right )\) [26, 32]. Note that in the MI literature this quantity is referred to as the microstructure function [33].
Spatial correlations between microstructure states can be quantified through the use of spatial statistics [24, 34]. The simplest of the n-point spatial statistics is two-point statistics. These quantities capture spatial correlations by considering the vector distance between two points. The example posed earlier in this section used two-point statistics to qualitatively describe the “spread” of crystals. Formally, the two-point statistics can be described by a conditional probability as follows:
where t is the vector that separates two points in the microstructure, g is the microstructure state at the tail of the vector, and \(\boldsymbol {g}^{\prime }\) is the microstructure state at the head of the vector. Note that if \(\boldsymbol {g}^{\prime }=\boldsymbol {g}\), then this quantity describes autocorrelation. Also, note that this quantity is solely a function of the difference in spatial location between two points (t); therefore, this definition assumes stationarity of the microstructure.
Consider now that we wish to obtain a compact representation of fx(g). There have been several works that have adopted the use of GSH for describing this quantity in polycrystalline systems [26, 27, 32]. Using a GSH basis fx(g) can be rewritten as follows:
where μ, n, l represent multiple indices for multiple sums, and \(F_{l\boldsymbol {x}}^{\mu n}\) is the complex-valued GSH coefficient at x which corresponds to the complex-valued GSH basis \(\dot {\dot {T}}_{l}^{\mu n}\). Note that the \(\dot {\dot {T}}_{l}^{\mu n}\) preserve crystal symmetries and are orthogonal to their complex conjugate \(\dot {\dot {T}}_{l}^{\mu n *}\). The coefficients \(F_{l\boldsymbol {x}}^{\mu n}\) can be obtained in the analogous way to how Fourier coefficients are determined (i.e. by exploiting orthogonality) \(F_{l\boldsymbol {x}}^{\mu n} = (2l + 1) \int f_{\boldsymbol {x}}\left (\boldsymbol {g}\right ) \dot {\dot {T}}_{l}^{\mu n*}\left (\boldsymbol {g}\right ) d\boldsymbol {g}\). In the case where spatial bin x is occupied by a single orientation go (an individual pixel in an indexed EBSD scan), then \(F_{l\boldsymbol {x}}^{\mu n} = (2l + 1) \dot {\dot {T}}_{l}^{\mu n*}\left (\boldsymbol {g}_{o}\right )\).
Naturally, the next step is to redefine the two-point statistics using the GSH basis representation. One practical consideration is that there are an infinite number of g to chose from since it is a continuous function. In recent works, this problem is overcome by computing spatial statistics over the complex-valued GSH coefficients themselves [26, 27]. The interpretation is that the different microstructure states are described by the different GSH coefficients indexed over μ, n, l. However, in this work, we will introduce one additional definition which produces a different interpretation of the spatial statistics. Here, we define an averaged quantity for the spatial autocorrelation, which averages over all g. In doing so, information about texture is lost, but this new definition is well suited for capturing the local misorientation or local morphological spatial behavior. Therefore, we define as follows:
where VFZ is the fundamental zone volume. Again, this quantity describes the spatial autocorrelation of crystallographic orientation averaged over all possible crystal orientations. Some information (texture) is lost but the structural morphological information is retained. The advantage of adopting this strategy is that often very large scans are needed to capture texture, which is inherently a volume-averaged quantity. Therefore, texture requires a large representative volume element (RVE) to be statistically representative of the material as a whole. Conversely, local morphological features may be representative at much smaller RVE length scales.
Combining the GSH representation of Eq. 2, definition in Eq. 3, and two-point statistics in Eq. 1, the following expressions may be derived as follows:
where \(f^{*}_{\boldsymbol {x}+\boldsymbol {t}}\left (\boldsymbol {g}\right )\) is the complex conjugate. Since f is a real valued function then f = f∗. This trick enables significant simplification when computing the product of the two large sums since we are using an orthogonal basis; \({\int }_{FZ} \dot {\dot {T}}_{l}^{\mu n} \dot {\dot {T}}_{l^{\prime }}^{\mu ^{\prime } n^{\prime } *} d\boldsymbol {g} = \left (2l + 1\right )^{-1}\) if all the indices “match” else 0. A similar manipulation was found in [32], but in their case, it was for computing localization relationships and not spatial autocorrelations. In fact, the definition introduced in Eq. 3 was purposefully introduced to exploit the orthogonality found in the GSH basis similar to what was done in [32]. This simplification only works for the case of autocorrelation; the orthogonality cannot be exploited when considering cross-correlations. The final expression obtained is a function (mean) of the autocorrelation statistics derived in [26, 27]. However, our derivation can be justified with some novel physical interpretation (mean autocorrelation over all g).
Note that although fx(g) is described using a truncated GSH expansion each of the GSH coefficients themselves is a complex-valued continuous variable. This treatment allows for gradations of similarity between pixels. For instance, pixels misoriented by only a few degrees will yield higher autocorrelation than pixels with large misorientation. If instead the continuous-valued microstructure state (orientation g) was discritized using a “binning” strategy [35], then pixels with similar orientations that happen to fall into different bins would erroneously suggest a lack of autocorrelation. Furthermore, binning of the three-dimensional orientation space would be cumbersome and inefficient [32].
The final line of Eq. 4 discretizes the spatial domain over \(\mathcal {X}\) into a two-dimensional-binned spatial domain over \(\mathcal {S}\) which corresponds to the EBSD scan pixels. The final expression is a convolution over S, which can be efficiently computed using discrete Fourier transforms (DFTs) [24]. The quantity |S| is the total number of spatial bins considered, e.g., total number of pixels in a image. Note for partial scans, scans where a portion of the image contains unreliable or “bad” measurements, recent algorithms have been established that account for this complication by modification of Eq. 4 [36].
The proposed microstructure descriptor is sensitive to the degree of GSH discretization introduced in Eq. 2. If too few terms are used in the sum, then it may be possible that fx(g) will be unable to accurately describe certain orientations present in the observed micrographs. Consequently, the morphologies associated with those inadequately resolved orientations will be neglected in the mean autocorrelation function (4). There are two recent works which address the question of GSH truncation when quantifying spatial microstructure data. Paulson et al. published a work on the homogenization of elastic and inelastic properties of polycrystalline HCP systems using a similar MI approach [26]. For HCP systems l = (0,2) yields 6 terms and l = (0,2,4) yields 15 terms in Eq. 2. In their study, they considered various crystallographic textures and found that truncation at 15 terms yielded marginally better results than at 6 terms. Yabansu, Patel, and Kalidindi found that truncation with l = (0,4), yielding a total of 10 terms, was suitable for building reduced-order elastic localization relationships in polycrystalline FCC systems [32]. Therefore, since there is evidence that both localization and homogenization relationships can be captured with minimal terms, we argue that a ten term GSH truncation should be sufficient for adequately describing the microstructures studied in this work.
Feature Selection and Bootstrapping
The previous section describes a rigorous method for quantifying the microstructure. The mean two-point statistics, \(\bar {p}_{\boldsymbol {t}}\), derived however is of the same dimensionality as t. Correspondingly, t is a vector that can be placed into the microstructure; hence, in this case, it is bounded by the size of the EBSD scans/images. Therefore, \(\bar {p}_{\boldsymbol {t}} \in \mathcal {R}^{N\times M}\) where N and M are the height and width of the images measured in pixels. All EBSD scans in this work are square; hence, the dimensionality of each statistic derived from the images is N2. Therefore, it is clear that for interpretability of the results some dimensionality reduction will be necessary. In this work, we utilize unsupervised principal component analysis (PCA), which computes a statistically optimal basis for describing the full feature space. PCA has been employed successfully in many MI works for compact representation of microstructure statistics [24, 26, 27, 37,38,39,40,41,42]. Dimensionality reduction is achieved by suitably truncating the basis expansion and using the basis weights (PC weights) to describe the data. This is analogous to Fourier representation of a one-dimensional signal where the Fourier coefficients can compactly describe the signal.
Another consideration when constructing the microstructure feature space is the need to ensure rotational invariance of the images. Consider that small deviations in how the samples are mounted in the microscope may result in angular rotation of the images, which therefore affects the microstructure statistics. Looking ahead at Fig. 8, careful inspection reveals that the statistics are slightly rotationally misoriented relative to one another. Failure to capture this experimental artifact could result in falsely discriminating two otherwise statistically identical microstructures. Rotational invariance is introduced by utilizing the methods found in [37]. Full details of this method are found in the referenced work and are not reproduced here.
Finally, a strategy is needed to obtain measurements of the dispersion of the PC weights. A naive and experimentally costly strategy would require that multiple EBSD scans be taken. From the dispersion (variance, covariance) measures, hypothesis testing could be performed or data-driven models could be built. This approach would be extremely expensive as each single scan is costly to obtain. An alternative strategy is to use the single observations and obtain dispersion estimates from bootstrapping of the images [43]. A similar strategy was utilized in [44, 45] for generating computationally efficient statistical volume elements (SVEs). Niezgoda, Yabansu, and Kalidindi utilized bootstrapping to obtain estimates of the structural variance of three-dimensional-simulated microstructures [46].
Bootstrapping seeks to establish dispersion estimates for mean quantities by a resampling of the data [43]. It is appealing because no distributional assumptions are needed (e.g., normality). Furthermore, it can be used to obtain dispersion estimates for complicated functions of the observed data. Consider that we make N observations of a normally distributed quantity X but we want the mean and mean-variance of some complicated function f(X).
In our setting, the data are the EBSD scans and the transformation is the pipeline that transforms the EBSD scans to \(\bar {p}_{\boldsymbol {t}}\) and then to the truncated PC weights. Special care is also needed to preserve the spatial correlation structure present in our data. Therefore, we used a strategy that is analogous to bootstrapping time-series data [43]: 7.5 μ m × 7.5 μ m images were sub-sampled four times and used to reconstruct a tiled 15 μ m × 15 μ m image. The 7.5 μ m × 7.5 μ m tiles were obtained by randomly selecting pixels from the image and then obtaining 3.75 μ m worth of pixels left, right, above, and below the selected point. In the case where the randomly selected pixel was within 3.75 μ m of a boundary the sub-sampled image was obtained by “wrapping” around the original image. For computing spatial statistics, this is acceptable since the convolution in Eq. 4 assumes periodic boundary conditions, which is equivalent to assuming that the image “wraps” around itself. This is shown schematically in Fig. 5. Each resampled 15 μ m × 15 μ m image corresponds to a single bootstrapped sample. For each setting, 100 bootstrapped samples were generated. The entire ensemble was then utilized to establish a PC basis and the corresponding PC weights for each bootstrapped sample were determined. The mean and variance of these bootstrapped PC weights were utilized to establish the mean and mean-dispersion at each unique process setting.
Multiple Output Gaussian Process Regression
A data-driven model is needed to efficiently map the controllable process parameters to the material quantities of interest. In this setting, the structure behaves as an intermediate variable that fundamentally controls the physics and is responsible for the exhibited properties. A statistical interpretation is that the structure variable is a latent variable; it is critically important but is either not possible to observe or perhaps can only be observed with great effort. This is an important consideration when identifying the relevant length scales and corresponding salient microstructural features. For instance, consider that TEM micrographs are rich with information at the lowest length scales but are costly to obtain. Conversely, optical micrographs are relatively easy to obtain but may have limited utility for certain problems, for instance, properties that are dependent on the lower length scale physics. Process-property models can sometimes capture the underlying relationships [47]; however, inclusion of structure into the modeling pipeline is preferred [24]. The justification is that structure physically governs the underlying property behavior and inclusion of such information may alleviate potential ambiguities associated with non-unique process-property mappings.
Modeling of PSP relationships traditionally follows a sequential strategy where the process-structure (PS) and structure-property (SP) relationships are established independently of one another and combined in sequence [40]. This is illustrated in the top of Fig. 6. A difficultly associated with such a framework is that it is not straightforward to quantify uncertainty propagation between PS and SP models. The PS model accepts process parameters as inputs, which are considered to be deterministic. The output microstructure estimates are naturally stochastic since the microstructure observations are stochastic. Computing confidence bounds for the output microstructure estimates is trivial in most statistical frameworks. Additional care however is needed in the subsequent SP modeling step when transferring forward stochastic structure estimates. As was just argued, the PS outputs are stochastic; hence, the SP inputs are stochastic. However, most data-driven models assume the model inputs to be deterministic.
Schematic of two modeling strategies for establishing PSP linkages. Note that italicized P refers to the process and normal font P represents properties. a A sequential strategy where process-structure and structure-property models are built independently and predictions flow sequentially, b jointly developed model using multiple output GP structure, which captures possible cross-correlations in the structure-property structure
Another limitation of the sequential PSP modeling strategy is that information is not shared across the PS and SP models. Consider that the model of interest is actually the full PSP model. This model is of course built using the two PS and SP submodels, which are usually established independently. A better PSP model could perhaps be established if the PS and SP models were built concurrently or perhaps with iteration; the best PS and SP models established independently may not produce the best PSP model.
GPR is a nonparametric curve fitting technique [48, 49]. Unlike traditional linear and nonlinear regression, nonparametric methods do not require a priori knowledge of the trends’ functional form. Instead, the data is assumed to come from a Gaussian data generating process where observations, yi and yj, may be correlated based on their proximity to each other, xi −xj. Future predictions, y(x), can be shown to be a weighted average of all the \(\left [y_{1},\ldots ,y_{N}\right ]\) where the weights depend on the proximity of x relative to all observations in the dataset \(\left [\boldsymbol {x}_{1},\ldots , \boldsymbol {x}_{N}\right ]\). The final form of the GPR statistical model is closely related to kernel regression and smoothing methods [48, 50].
In classical regression, the statistical inference or learning is performed by optimally estimating the unknown regression coefficients. In the GPR setting, the inference is performed by estimating the unknown statistical hyperparameters. These quantities define the correlation structure, which is embedded in the collected observations. For instance, correlation length scales are used to precisely quantify the relative measures of “proximity” mentioned above. In some problems, xi − xj = 1 may be an insignificant difference; yet in other instances, this may be large.
In this work, we attempt to address both these considerations by utilizing a multiple output Gaussian process regression (MOGPR) model for simultaneously identifying the full PSP model (Fig. 6). Multiple output implies that y need not be a scalar. This model choice offers several promising features not available using a sequential strategy. Firstly, structure and properties are modeled together as a function of process inputs using a multivariate normal structure to quantify structure-property correlations. In this way, process-property is possible; however, the model will also infer possible SP correlations when present. The SP cross-correlation jointly considers the full PSP linkage rather than independent submodels. Secondly, quantifying the SP variables simultaneously in a multiple output setting allows for easy uncertainty quantification of all relevant quantities including their cross-correlation structure. Finally, the MOGPR framework is flexible in its treatment of data and enables the inclusion of partial datasets with missing data. For instance consider a study where there are two microstructure descriptors. One is obtained using efficient experiments such as optical microscopy. The other descriptor is obtained using TEM and is therefore costly to acquire. The dataset may therefore contain many times more optical images than TEM images. However, in establishing the SP linkages standard regression models require both covariates for each individual property measurement. Clearly, such a framework cannot pair the two descriptors since one is much more numerous! The state of the art in this setting is to implement a transfer learning model which enables sharing of information between the two kinds of structure data [51]. The MOGPR model can automatically accommodate this setup. Additional details on the GPR framework, estimation of hyperparameters, prediction estimates, and details on the implementation used in this work are found in Appendix A.
Multi-Fidelity Property Modeling
In this work, structural descriptors come from the PC weights of the mean crystallographic autocorrelation function (\(\hat {p}_{\boldsymbol {t}}\)). Property measurements are obtained using spherical nanoindentation. The indentation stress-strain yield strength is used to quantify material strength [29]. The fraction explained variance (Fig. 7) illustrates that two PC components capture 97% of the observed variance. Therefore, in this study, M = 3 where j = 1,2 are the first two PC weights and j = 3 is the indentation yield, e.g., the MOGPR model represents the vector \(\left (PC_{1},PC_{2},Y_{\text {ind}}\right )\). Yind has some physically meaningful interpretation but is a somewhat noisy observation (see, Fig. 16). This variation is inherited from various sources including microstructure and surface characteristics. In Figs. 3 and 4, it is clear that the indenter could possibly engage different crystallographic orientations from test to test. Furthermore, there is also morphological heterogeneity across microstructures. Although the final contact radius using a 100 μ m indenter is on the order of 10–20 μ m, the contact radius is at the yield point roughly 1–2 μ m. Even using a larger 500 μ m indenter would not produce RVEs of crystallographic orientation and larger indenters (the next available indenter is 1500 μ m) are not feasible due to the load limits of the machine and the size of our samples (the smallest is 500 μ m in thickness). A brute-force strategy would require EBSD imaging of every SVE indentation site, which is experimentally costly. Finally, the response is sensitive to nanoscale asperities on the prepared surfaces, which introduces variation in the form of noise.
Therefore, in this work, our strategy is to simply homogenize over these effects; therefore, we have conducted many repeated indentation experiments for each unique process setting. However, a complimentary strategy is available that allows the combination of nanoindentation data with cheaper lower fidelity property data. In the statistics community, this is referred to as multi-fidelity modeling [52,53,54]. For this work, we consider the Vickers microhardness (HV) as a cheap property measure. The justification is that the spherical indentation stress-strain protocols enable granular interpretation of both elastic and post-elastic behavior of the indented material whereas hardness does not. Nevertheless, microhardness is easy to obtain and therefore may aid in bolstering confidence in our inferences. Additionally, the hardness data is less noisy because it is less sensitive to the previously described heterogeneities since the volume of material probed is much larger; diagonals produced during indentation at a 500 g of load were on the order of 80–100 μ m. A key assumption here is that Yind and HV follow the same trends. We will introduce some flexibility, however, in case they do not follow the same trends or if they do not follow the same trends under certain process settings. The necessary statistical framework for incorporation of multi-fidelity property data may be found in Appendix B.
Results
The mean crystallographic autocorrelation for each micrograph is shown in Fig. 8. Note that these autocorrelation statistics are empirical quantities as they are computed directly from the data using Eq. 4, which is free from any parametric assumptions. It is important to acknowledge this as subsequent modeling is performed by directly comparing these statistics and therefore the same field of view (FOV) must always be used. All the statistics shown in Fig. 8 have a field of view of 15 μ m. Therefore, images obtained at α = 25∘,45∘, which have FOV of 45 and 105 μ m, were subsampled. The analysis therefore does not consider autocorrelation information available at larger correlation lengths in these images. However, this “clipping” is necessary to maintain identical scales across all the empirically computed autocorrelations.
Bootstrapped samples of the rotationally invariant mean crystallographic autocorrelation are shown in Fig. 9. Recall that 97% of the variance can be captured with a truncated PCA expansion using only two principal components (see, Fig. 7). Also shown in Fig. 9 is the predicted MOGPR path in PC space. The bootstrapped samples visually appear to generate scatter close to a bivariate normal distribution. Both the degree of scatter and the correlation in the scatter varies for each unique process setting. Therefore, the components of the observation error covariance matrix, Σ, which correspond to these structural variables were prescribed using frequent estimates for each unique process setting. This simplification is justified since the scope of our work is to quantify and model mean quantities. Additionally, bootstrapping is an effective method for estimating the dispersion of statistics; therefore, the hyperparameter inference in Eq. A.8 can be simplified. Furthermore, since the repetitions themselves only capture dispersion information of the data, and the observation error is specified, it is only necessary to utilize the mean value structure variables, \(\bar {\boldsymbol {PC}}_{i}\), when building the MOGPR model. This final point saves a great deal of computational burden associated with inverting C + Σ. This simplification requires only 16 two-dimensional mean values rather than the full data set.
In Figs. 10 and 11, the structure-property relations are shown. Note that SP data are not paired; there is not a “corresponding” property measure for each micrograph. Visualization, however, requires pairing; therefore, the mean values and the associated confidence intervals are shown for experimental data. The mean MOGPR path and the confidence region are also shown. Note that there is a clear distinction between the confidence region of the mean and confidence region of future observations. Future observations will also contain some observation errors and would therefore have a correspondingly larger confidence region. At V = 1.00m ⋅s− 1, the trends appear to change despite the behavior being fairly consistent across cutting speeds V < 1.00m ⋅s− 1. This experimental setting corresponds to the largest imposed temperatures since \({\Delta } t \sim 1/V\); hence, there is less time available for conduction of heat away from the generated chips [55].
PS relationships are shown in Figs. 12, 13 and 15. It is clear that the rake angle, α, has the greatest influence on the generated structures. This agrees with intuition as α controls the geometric configuration of the experiment, and therefore has the greatest impact on the imposed shear strains γ. Deformation conversely drives structural refinement and evolution via the DRX mechanism [3].
Finally, the process-property maps are shown in Fig. 16. Note that process-property implicitly considers structural relationships via the MOGPR model. The Vickers hardness data generally follows trends similar to the indentation yield. At the highest cutting speed, V = 1.00m ⋅s− 1, there is a significant decrease in hardness/strength going from α = 15∘ to α = 5∘. This is only observed at the highest speed, which suggests that physically this anomalous behavior is driven by thermal effects.
Discussion
The proposed mean crystallographic autocorrelation spatial statistic is an effective measure of microstructural morphology. The power of this metric is that it quantifies morphology without the need to explicitly define microstructural features. A common assumption when analyzing EBSD data is to define a threshold misorientation value for defining high angle boundaries. At other times, the misorientation distribution function (ODF) itself is utilized as a metric, but this necessitates identification of grain boundaries, which is again based on assumed threshold values [5]. Since our statistic only captures morphological features, it may be well suited in settings where the scan size is smaller than what is required for accurately quantifying texture. Crystallographic texture is a homogenized quantity; therefore, larger scans are typically necessary to accurately capture the representative crystallographic texture. The 15μ m × 15μ m images in Fig. 4 are certainly not sufficient for identifying texture but can still be used for quantifying morphological features.
Physical interpretation of the obtained microstructure evolution results is possible by considering the PCA bases shown in Fig. 7. Recall that \(\bar {p}_{\boldsymbol {t}}\) measures the degree of spatial crystallographic autocorrelation (similarity). The first principal basis corresponding to PC1 is highly localized with large negative values towards the center of the basis, some positive asymmetric values away from 𝜃 = 0∘, and slightly positive in the remainder of the region. The peaked negative region corresponds to a length of about 10 pixels which is 500nm (50nm/pixel). Note that this corresponds to the refined crystallite size observed at the largest strains. Conversely, PC2 has an even sharper, but faint, negative peak in the center, positive values in the 0.5 − 2μ m range, and negative values at large distances. Therefore, one contribution of the PC1 basis is to control a high autocorrelation region concentrated within a 500-nm region. PC2 captures competing autocorrelation trends in the 0.5 − 2μ m and > 3 μ m range. Therefore, it is reasonable that PC1 is observed to displays the greatest sensitivity to the applied rake angle (Figs. 12 and 13). As the rake angle is decreased, strains are increased, DRX drives refinement, and therefore pixels only retain autocorrelation with very close neighbor points (roughly within a crystal). However, PC1 does not appear to significantly change with cutting speed (Fig. 14). This is because cutting speed does not influence spatial similarity at these small scales. PC2, however, does appear to be sensitive to cutting speed (Fig. 15), and this sensitivity decreases with increasing rake angle (decreasing strain). This implies that at large imposed strains, as the cutting speed is increased, similarity of crystal orientation extends to include larger neighborhoods in the 0.5 − 2μ m region. This observation agrees with the process physics where it is known that cutting temperatures increase with both increasing speeds and strains. Additional straining drives heat generation via plastic dissipation and increased cutting speeds limit the efficacy of conduction to remove heat away from the process zone. At higher temperatures, DRX is less impactful [3]; thus, there is less misorientation and hence crystal similarity extends over larger spatial distances (less misorientation). Therefore, PC2 is sensitive to thermal effects, which are implicitly tied to the cutting speeds. With respect to the rake angle, PC2 has a significant quadratic interaction and this complex behavior may be explained as follows. At high rake angles (low strains), the similarity extends over large distances (> 3μ m) and PC2 is negative, which yields large positive autocorrelation values at large distances. With increasing strain (decreasing rake angle), there is less autocorrelation at large length scales but correlations in the intermediate values (0.5 − 2μ m) persist and hence PC2 increases. However, this trend reverses at the lowest rake angles (highest strains) when the autocorrelation becomes extremely localized (< 500 nm); thus, less similarity is observed in the 0.5 − 2μ m range. These interactions are complex because each basis captures several coupled physical features (e.g., PC2 captures negative long range and positive medium range autocorrelation). Furthermore, the bases must interact and balance their respective contributions in order to describe the changing physics at different machining process settings.
Figure 16 illustrates that the Vickers hardness and indentation yield produce similar trends with respect to the rake angle. For reference, the mean virgin material hardness is HV = 87.5 ± 5.0 (95% confidence interval). At large rake angles (low strains), the generated chips have higher hardness than the virgin material but produce lower range properties relative to measurements at small rake angles (larger strains). This observation is in accordance with deformation induced strain hardening. For cutting speeds V = 0.20,0.33,0.50m ⋅s− 1, the hardness appears to saturate with decreasing rake angle, which indicates that additional straining does not drive an increase in hardness. At the lowest cutting speed, however, indentation yield produces a fairly linear trend which decreased with increasing speed. Therefore, hardness and yield do not always share a one-to-one correspondence, but nevertheless, the inclusion of hardness is informative. At the highest speed and lowest rake angle (highest strain), there is a significant decrease in both hardness and strength. This is likely driven by recovery processes, which occur due to the higher cutting temperatures experienced under these conditions.
In this study, we only consider structural morphology and therefore neglect crystallographic effects. This is one potential source of the scatter observed in Fig. 16. The local crystallographic orientation of the indented site will likely influence the indentation response. However, for simplicity, we adopt a strategy where this was neglected and instead homogenized over many observations. When crystallographic information is desirable, the stand-alone GSH representation (which quantifies the ODF) may be augmented as additional features to \(\bar {p}_{\boldsymbol {t}}\). Another possibility is to use the strategy established in [26, 27] and use the paired two-point statistics between each of the GSH coefficients. Recall that the GSH representation is a sum over multiple indices (μ, n, l), and in this work, we truncate to 10 terms. Each of these 10 terms can be used as a measure of microstructural state. Therefore, these state descriptors may be used to compute two-point spatial correlations [26]. Including constraints and symmetry considerations, it may be shown that there are 2 ⋅ 10 − 1 = 19 unique correlation pairs [26]. The derived expression in Eq. 4 happens to be the mean over all autocorrelation pairs considered in [26]. The derivation in this work is fairly compact and proves that this mean quantity has a physical interpretation and is a descriptor of morphological spatial crystallographic “spread”, which includes misorientation.
Bootstrapping methodology appears to be an effective method for quantifying the dispersion of microstructure in reduced order PC space, as shown in Fig. 9. In our regression model, bootstrapping is useful as it eliminates the need to estimate the measurement error variances when training the MOGPR model—instead, they can be estimated directly from bootstrapping. Note, however, that bootstrapping of correlated data requires that the original sample be sufficiently large such that it “contains” the relevant correlation length scales. In our setting, the correlation length scales, particularly at large rake angles (low strain), are larger than the image field of view. Nevertheless, the bootstrapped variance estimates will reflect this artifact; inadequately sized images will yield more variance. Additionally, the disparity in autocorrelation at the lower spatial length scales is sufficiently significant that trends are still clear despite “missing” information at very large length scales.
The MOGPR model is effective at quantifying PSP relationships and provides estimates for coupled SP uncertainties. A natural concern, however, is that perhaps the obtained hyperparameters, \(\hat {\boldsymbol {\Phi }}\) in Eq. A.8, neglect SP relationships. In Eq. A.6, the SP linkage is captured through the cross-correlation matrix S, which must be inferred from the observed data. This matrix quantifies the covariance (or correlation) between all the outputs considered (PC1, PC2, Yind). The case where structure-property linkages are neglected the covariance matrix would take a block form as follows:
which suggests no correlation between the PC s and Yind. This degenerate case corresponds to two independent GP models; one for the PS and another for process-properties. Yet, another degenerate case corresponds to a diagonal covariance structure where no correlation exists between any of the considered variables; thus, the result is M independent GP models. However, consider that this model is data-driven; therefore, it is possible that perhaps a process-property relationship does exist. In fact, there is some recent evidence in the literature that suggests that these mappings are plausible in some settings [56]. The inclusion of structural information is physically motivated and is expected to yield better performance as the data is much richer if structure information is included. The merit of the MOGPR model is that all possibilities may be considered at once; if a direct process-property linkage exists then the model will identify it. Note that it may seem inappropriate to assume structure-structure cross-correlations between the PC weights as PCA theory generates PC weights which are independent, e.g., \(\text {Cov}\left (PC_{1},PC_{2}\right )= 0\). However, this is only true in the unsupervised setting; PC weights are independent when nothing is known about the process settings. The PC basis and weights are computed from the unlabeled pt ensemble of observations. In the MOGPR model, correlation between PC1 and PC2 is possible because the correlation is conditional on also knowing the process settings. Two uncorrelated random variables may become correlated when conditioned on a third random variable related to the first two. Clearly, in the second case, the two otherwise independent experiments become correlated due to the extra information.
Cross-validation results using a leave-one-unique-process-setting-out strategy are displayed in Fig. 17. The cross-validation results may be exactly computed from the fully trained model by employing a shortcut formula (see, Appendix C). Four different results are shown to illustrate a few key points: (a) cross-validation using Yind property data not including output cross-correlations (the case of M independent GP models); (b) cross-validation using Yind property data with SP cross-correlations; (c) cross-validation using Yind and HV as coupled properties with no SP cross-correlations; and finally, (d) cross-validation considering all available property data (Yind and HV ) and including output cross-correlations. Notice that strategy (d), which considers all property data and all correlations, yields the best cross-validation error (25% improvement in Yind prediction relative to model (a)). Therefore, inclusion of the hardness data did improve the overall model performance. Furthermore, each increase in model complexity provides slight improvements over the previous model. In general, however, this may not always be the case. GPR models are also prone to over-fitting when there is an imbalance between model complexity and data. For this reason, some researchers prefer to use cross-validation strategies for model training [57].
Conclusions
In this work, we studied a severe plastic deformation machining process which drives microstructure evolution via continuous dynamic recrystallization. Various stages of microstructure evolution were captured by considering a wide range of rake angles, which induce a wide range of shear strains. Rate and temperature effects were considered by varying the cutting speed. Large strain conditions produced submicron crystal structures, whereas low-strain experiments yielded highly deformed structures, which still resembled the coarse parent material. At the largest strains, a dependence on the cutting speed was observed with higher cutting speeds producing structures with lower crystallographic misorientations. Generalized spherical harmonics were used to efficiently quantify the local orientation state and a novel autocorrelation spatial statistic was derived that captures orientation “spread” or misorientation. The novel descriptor is physically intuitive and targets morphological information present in the orientation imaging data. A data-driven multiple output Gaussian process regression model was established for quantifying process-structure-property linkages. The model is flexible, enables inclusion of various kinds of structure and property data, does not necessitate fully paired input data, captures the full process-structure-property pipeline, and produces coupled uncertainty estimates associated with future predictions.
References
Shaw MC, Cookson J (1984) Metal cutting principles. Clarendon Press, Oxford
Sagapuram D, Yeung H, Guo Y, Mahato A, M’Saoubi R, Compton WD, Trumble KP, Chandrasekar S (2015) On control of flow instabilities in cutting of metals. CIRP Annals 64(1):49
Brown TL, Saldana C, Murthy TG, Mann JB, Guo Y, Allard LF, King AH, Compton WD, Trumble KP, Chandrasekar S (2009) A study of the interactive effects of strain, strain rate and temperature in severe plastic deformation of copper. Acta Materialia 57(18):5491–5500
Basu S, Shankar MR (2015) Crystallographic textures resulting from severe shear deformation in machining. Metall Mater Trans A 46(2):801–812
Wang Z, Basu S, Murthy TG, Saldana C (2018) Gradient microstructure and texture in wedge-based severe plastic burnishing of copper. J Mater Res 33(8):1046
Wang Z, Basu S, Saldana C (2017) Low-temperature machining in a fully submerged cryogenic environment. Mach Sci Technol 21(1):19
Guo Y, Saldana C, Compton WD, Chandrasekar S (2011) Controlling deformation and microstructure on machined surfaces. Acta materialia 59(11):4538–4547
M’Saoubi R, Larsson T, Outeiro J, Guo Y, Suslov S, Saldana C, Chandrasekar S (2012) Surface integrity analysis of machined inconel 718 over multiple length scales. CIRP Ann-Manuf Technol 61(1):99–102
Ni H, Elmadagli M, Alpas A (2004) Mechanical properties and microstructures of 1100 aluminum subjected to dry machining. Mater Sci Eng A 385(1-2):267–278
Swaminathan S, Shankar M, Lee S, Hwang J, King AH, Kezar RF, Rao BC, Brown TL, Chandrasekar S, Compton WD et al (2005) Large strain deformation and ultra-fine grained materials by machining. Mater Sci Eng A 410:358
Valiev RZ, Islamgaliev RK, Alexandrov IV (2000) Bulk nanostructured materials from severe plastic deformation. Progress Mater Sci 45(2):103–189
Zhilyaev AP, Langdon TG (2008) Using high-pressure torsion for metal processing: Fun- damentals and applications. Prog Mater Sci 53(6):893–979
Murr L, Ramirez A, Gaytan S, Lopez M, Martinez E, Hernandez D, Martinez E (2009) Microstructure evolution associated with adiabatic shear bands and shear band failure in ballistic plug formation in ti-6al-4v targets. Mater Sci Eng A 516(1–2):205–216
Minnaar K, Zhou M (1998) An analysis of the dynamic shear failure resistance of structural metals. J Mech Phys Solids 46(10):2155–2170
Me-Bar Y, Shechtman D (1983) On the adiabatic shear of ti 6al 4v ballistic targets. Mater Sci Eng 58 (2):181–188
Fatemi-Varzaneh S, Zarei-Hanzaki A, Beladi H (2007) Dynamic recrystallization in az31 magnesium alloy. Mater Sci Eng A 456(1–2):52–57
Ion S, Humphreys F, White S (1982) Dynamic recrystallisation and the development of microstructure during the high temperature deformation of magnesium. Acta Metall 30(10):1909–1919
Tóth L, Beausir B, Gu C, Estrin Y, Scheerbaum N, Davies C (2010) Effect of grain refinement by severe plastic deformation on the next-neighbor misorientation distribution. Acta Mater 58(20):6706–6716
Abolghasem S, Basu S, Shekhar S, Cai J, Shankar M (2012) Mapping subgrain sizes resulting from severe simple shear deformation. Acta Mater 60(1):376–386
Shekhar S, Abolghasem S, Basu S, Cai J, Shankar M (2012) Effect of severe plastic deformation in machining elucidated via rate-strain-microstructure mappings. J Manuf Sci Eng 134(3):031008
Basu S, Wang Z, Liu R, Saldana C (2016) Enhanced subsurface grain refinement during transient shear-based surface generation. Acta Mater 116:114–123
Kalidindi SR, Medford AJ, McDowell DL (2016) Vision for data and informatics in the future materials innovation ecosystem. JOM 68(8):2126–2137
Kalidindi SR, Brough DB, Li S, Cecen A, Blekh AL, Congo FYP, Campbell C (2016) Role of materials data science and informatics in accelerated materials innovation. Mrs Bull 41(8):596–602
Kalidindi SR (2015) Hierarchical materials informatics: novel analytics for materials data. Elsevier
Niezgoda SR (2010) Stochastic representation of microstructure via higher-order statistics: theory and application
Paulson NH, Priddy MW, McDowell DL, Kalidindi SR (2017) Reduced-order structure property linkages for polycrystalline microstructures based on 2-point statistics. Acta Mater 129:428–438
Paulson NH, Priddy MW, McDowell DL, Kalidindi SR (2018) Materials & Design
Yang Z, Li X, Brinson LC, Choudhary AN, Chen W, Agrawal A (2018) arXiv:1805.02791
Pathak S, Shaffer J, Kalidindi SR (2009) Determination of an effective zero-point and extraction of indentation stress-strain curves without the continuous stiffness measurement signal. Scr Mater 60(6):439–442
Pathak S, Stojakovic D, Doherty R, Kalidindi S (2009) Importance of surface preparation on the nano-indentation stress-strain curves measured in metals. J Mater Res 24(3):1142–1155
Bunge HJ (2013) Texture analysis in materials science: mathematical methods. Elsevier
Yabansu YC, Patel DK, Kalidindi S (2014) Calibrated localization relationships for elastic response of polycrystalline aggregates. Acta Mater 81:151–160
Adams BL, Gao XC, Kalidindi S (2005) Finite approximations to the second-order properties closure in single phase polycrystals. Acta Mater 53(13):3563–3577
Torquato S (2013) Random heterogeneous materials: microstructure and macroscopic properties, vol 16. Springer Science & Business Media
Fast T, Niezgoda SR, Kalidindi S (2011) A new framework for computationally efficient structure-structure evolution linkages to facilitate high-fidelity scale bridging in multiscale materials models. Acta Mater 59(2):699–707
Cecen A, Fast T, Kalidindi SR (2016) Versatile algorithms for the computation of 2-point spatial correlations in quantifying material structure. Integr Mater Manuf Innov 5(1):1
Cecen A, Yabansu YC, Kalidindi SR (2018) A new framework for rotationally invariant two-point spatial correlations in microstructure datasets. Acta Materialia
Cecen A, Fast T, Kumbur E, Kalidindi S (2014) A data-driven approach to establishing microstructure-property relationships in porous transport layers of polymer electrolyte fuel cells. J Power Sources 245:144–153
Iskakov A, Yabansu YC, Rajagopalan S, Kapustina A, Kalidindi S (2018) Application of spherical indentation and the materials knowledge system framework to establishing microstructure-yield strength linkages from carbon steel scoops excised from high-temperature exposed components. Acta Mater 144:758–767
Khosravani A, Cecen A, Kalidindi SR (2017) Development of high throughput assays for establishing process-structure-property linkages in multiphase polycrystalline metals: Application to dual-phase steels. Acta Mater 123:55–69
Sundararaghavan V, Zabaras N (2004) A dynamic material library for the representation of single-phase polyhedral microstructures. Acta Materialia 52(14):4111–4119
Sundararaghavan V, Zabaras N (2005) Classification and reconstruction of three-dimensional microstructures using support vector machines. Comput Mater Sci 32(2):223–239
Efron B, Tibshirani RJ (1994) An introduction to the bootstrap. CRC Press
Wargo E, Hanna A, Cecen A, Kalidindi S, Kumbur E (2012) Selection of representative volume elements for pore-scale analysis of transport in fuel cell materials. J Power Sources 197:168–179
Cecen A, Wargo E, Hanna A, Turner D, Kalidindi S, Kumbur E (2012) 3-d microstructure analysis of fuel cell materials: spatial distributions of tortuosity, void size and diffusivity. J Electrochem Soc 159 (3):B299–B307
Niezgoda SR, Yabansu YC, Kalidindi S (2011) Understanding and visualizing microstructure and microstructure variance as a stochastic process. Acta Mater 59(16):6387–6400
Deshpande P, Gautham B, Cecen A, Kalidindi S, Agrawal A, Choudhary A (2013) Application of statistical and machine learning techniques for correlating properties to composition and manufacturing processes of steels. In: Proceedings of the 2nd World congress on integrated computational materials engineering (ICME). Springer, pp 155–160
Rasmussen CE (2004) In: Advanced lectures on machine learning. Springer, pp 63–71
Santner TJ, Williams BJ, Notz WI (2013) The design and analysis of computer experiments. Springer Science & Business Media
Bishop CM (2006) Pattern recognition and machine learning (information science and statistics). Springer, Berlin
Pan SJ, Yang Q, et al. (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Haaland B, Qian PZ (2010) An approach to constructing nested space-filling designs for multi-fidelity computer experiments. Stat Sin 20(3):1063
Tuo R, Wu CJ, Yu D (2014) Surrogate modeling of computer experiments with different mesh densities. Technometrics 56(3):372–380
Kennedy MC, O’Hagan A (2001) Bayesian calibration of computer models. J R Stat Soc Ser B (Stat Methodol) 63(3):425–464
Shaw MC, Cookson J (2005) Metal cutting principles, vol 2. Oxford University Press, New York
Agrawal A, Deshpande PD, Cecen A, Basavarsu GP, Choudhary AN, Kalidindi SR (2014) Exploration of data science techniques to predict fatigue strength of steel from composition and processing parameters. Integr Mater Manuf Innov 3(1):8
Pilania G, Mannodi-Kanakkithodi A, Uberuaga B, Ramprasad R, Gubernatis J, Lookman T (2016) Machine learning bandgaps of double perovskites. Sci Rep 6:19375
Fuentes M (2001) A high frequency kriging approach for non-stationary environmental processes. Environmetrics: Off J Int Environmetr Soc 12(5):469–483
Boyle P, Frean M (2005) Dependent gaussian processes. In: Advances in neural information processing systems, pp 217–224
Fernandez-Zelaia P, Melkote SN (2018) Statistical calibration and uncertainty quantification of complex machining computer models. International Journal of Machine Tools and Manufacture
Fernandez-Zelaia P (2019) Machining psp. https://github.com/pfz3
Fernandez-Zelaia P, Joseph VR, Kalidindi S, Melkote SN (2018) Estimating mechanical properties from spherical indentation using bayesian approaches. Mater Des 147:92–105
Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, Brubaker M, Guo J, Li P, Riddell A (2017) Stan: A probabilistic programming language. J Statist Softw, 76(1)
Stan Development Team (2018) RStan: the R interface to Stan. http://mc-stan.org/. R package version 2.17.3
Hoff PD (2009) A first course in Bayesian statistical methods. Springer Science & Business Media
Acknowledgements
The authors are grateful to the Woodruff School machine shop for their assistance in manufacturing of the custom cutting tool used in this work.
Funding
Financial support of the work by the Morris M. Bryan, Jr. Professorship is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: MOGPR Implementation
Consider a process who’s input are x and that has multiple outputs \(\left [Y_{1},Y_{2},\ldots , Y_{K}\right ]\) which are observed with some measurement error 𝜖. This process can be modeled using a multivariate GP model as follows:
where the mean behavior of the outputs varies according to a mean function\(\boldsymbol {\mu }\left (\boldsymbol {x}\right )\), cross-correlation of outputs are captured through S, and the observation errors are in general allowed to be correlated and perhaps have different scales for each outputs, e.g., Cov𝜖 is purposefully generic. The mean function may be described using a parametric regression strategy, e.g., μ(x) = f(x)β where β are regression coefficients and f(x) a vector of regressors. In this work, we utilized linear and cross-linear terms (4 terms including constant) for each output Yi. Note that we implicitly assume that the cross-correlations are spatially invariant and therefore stationary. Strategies exist for introducing nonstationarity [58, 59] and we have successfully utilized these for developing FE surrogates; however, in this work, we will utilize the simpler stationary cross-correlation structure [60]. The codes provided online, however, include additional nonstationary complexity [61].
Now consider that observations of each output Yi are made at xij where j = 1,…, Ni. This notation is flexible enough to allow each Yi output to have Ni unique observations xij with a total number of \(N={\sum }_{i = 1}^{K} N_{i}\). Again, this is valuable if the Yi have different costs associated with obtaining them (optical vs. TEM). The accumulated dataset therefore can be expressed as another multivariate normal as follows:
where R is the Gaussian correlation function, Σ is the total error covariance matrix, and ϕi are the correlation length scales for each of the d dimensions of x. Note that Σ is comprised of σij and contains some flexibility for different kinds of experiments. Observations have some measurement variance σij when outputs are identical (i = j) and are observed at the same x (k = l); however, if there are paired observations (Yi, Yj) at each observation (k = l), then there may also be correlations in the errors. One example where this may be relevant is when considering the PC weights as microstructure descriptors which will generate pairs (or tuples in higher dimensions) of data for each micrograph. If observations are not measured in pairs, then they should be independent and share no correlation. Note that if all experiments are performed at the same x then the above covariance structure has a Kronecker structure which can be exploited for computational efficiency [62]. The covariance structure imposes that there is a distance-based criteria for quantifying correlations (R), that there are cross-correlations across outputs (Σ), and that there is a random component associated with measurement uncertainty (Σ).
The prior placed on the data is that observations can be explained by interpreting them as coming from some multivariate normal generating process. As such the unknown hyperparameters Φ = [β, ϕ, S, Σ] can be obtained from a maximum likelihood estimate (MLE) or a maximum a posteriori (MAP) estimate if priors are placed on some hyperparameters. The negative log-likelihood (or in a Bayesian setting the posterior) may be minimized to obtain estimates as follows:
The statistical model was built in Stan [63], a statistical programming language, and evaluated using RStan [64] the R-language interface for Stan. The Stan optimizer was used to identify the MAP estimates of the hyperparameters \(\hat {\boldsymbol {\Phi }}\). Note that in the inference there are two matrix quantities that need to be estimated. To improve efficiency of the estimation and maintain that these matrices remain positive definite inverse-Wishart priors were placed on the matrix quantities and the inference was reparameterized. A review of this strategy can be found in [65].
Note that in Section 1, a methodology for bootstrapping confidence bounds on the mean PC weights is presented. Therefore, for each unique process setting, we can establish the mean and variance measures of the mean PC weights. The mean estimates should be used in constructing the data vector, Y, and the bootstrapped variance estimates can be prescribed to build the structure-portion of the error covariance matrix Σ. This methodology also allows for inclusion of heteroskedastic variance estimates, which according to Fig. 9, are appropriate. Furthermore, this rearrangement alleviates some computational burden associated with estimating some hyperparameters and enables use of bootstrapped quantities.
Predictions using a tuned MOGPR model can be easily obtained by again considering that the desired quantities, \(\left (Y_{1}(\boldsymbol {x}),\ldots ,Y_{M}(\boldsymbol {x})\right )^{T}\), are jointly distributed with the observed data as follows:
where r captures the spatial and cross-correlations of each output Yi(x) with all previously observed data j = 1,…, M. The expectation of \(\left (Y_{1}(\boldsymbol {x}),\ldots ,Y_{M}(\boldsymbol {x})\right )^{T}\) conditional on all previous observations \(\left (\boldsymbol {Y}_{1},\ldots ,\boldsymbol {Y}_{M}\right )^{T}\) is as follows:
and the covariance associated with this prediction is as follows:
From these two expressions, it is clear that the MOGPR provides estimates for the PSP linkages as well as uncertainty predictions through the prediction covariance \(\hat {\boldsymbol {S}}\).
Appendix B: Multi-fidelity Implementation
Statistically, we can build a simple model that allows for sharing of information between the physically informative quantities (Yind) and the cheaper less informative quantities (HV ) as follows:
where Z is the underlying mean function, we seek described as a Gaussian process (GP), τ is error associated with Yind, ρ is a scaling quantity, W is an independent zero-mean GP which allows HV to vary from ρZ systematically (e.g., bias function), and γ is the measurement error in HV. This form is identical to the form introduced in the seminal Kennedy and O’Hagan paper [54]. Note that Z is part of the multivariate GP previously introduced but we simply denote it here as Z for simplicity. The model states that HV scales with Z (and hence the mean of Yind) except when the simple scaling fails in which case W “captures” or “soaks up” this deviation.
The covariance of HV with the other MOGPR quantities can be easily derived. First assume that Yind = YM− 1 in the model, e.g., the indentation yield is ordered as the second to last output, and HV is the last YM = HV. Therefore,
where \({\sigma ^{2}_{b}}\) and Rb are the bias variance and correlation function. The bias correlation function contains additional hyperparameters ϕb. Note there is no error term since there are no off-diagonal terms in the error covariance structure. This is because indentation-hardness experiments are not “paired” experimentally; observations are made independently of one another. All other correlations can be easily obtained simply by following the above “plug-in” strategy. The bias function “kicks in” only for HV − HV covariances as follows:
Appendix C: Cross-Validation Shortcut Formulas
The cross-validation error associated with removing a subset of data of size Ni, represented by multi-index i, can be expressed as follows:
where yi are the responses corresponding to i and \(\hat {\boldsymbol {f}}_{(i)}(\boldsymbol {x}_{i})\) is the corresponding estimate for a model which is trained by withholding data belonging to i. Note that cvi is a vector with multiple observations of a potentially multivariate output (e.g., PC1, PC2, Yind). The model estimate is given by the following:
where quantities containing subscript (i) represent quantities computed with data from i missing.
Now consider the complete covariance matrix where the ordering of the data is rearranged such that the block corresponding to i is shifted to the final rows/columns as follows:
To compute C− 1, the Sherman-Morrison-Woodbury formula can be applied as follows:
where \(\boldsymbol {B}=\boldsymbol {C}_{(i)}^{-1}\widetilde {\boldsymbol {r}}_{(i)}\left (\widetilde {\boldsymbol {\Sigma }}_{i}-\widetilde {\boldsymbol {r}}_{(i)}^{T}\boldsymbol {C}_{(i)}^{-1}\widetilde {\boldsymbol {r}}_{(i)}\right )^{-1}\). Note that this manipulation enables the interpretation of quantity \(\left (\widetilde {\boldsymbol {\Sigma }}_{i}-\widetilde {\boldsymbol {r}}_{(i)}^{T}\boldsymbol {C}_{(i)}^{-1}\widetilde {\boldsymbol {r}}_{(i)}\right )^{-1}\) as the i th “block-diagonal” entry of C− 1. Here, we are referring to the Ni × Ni entry corresponding to indices i which will be noted as \(\boldsymbol {C}_{ii}^{-1}\). Similarly \(-\left (\widetilde {\boldsymbol {\Sigma }}_{i}-\widetilde {\boldsymbol {r}}_{(i)}^{T}\boldsymbol {C}_{(i)}^{-1}\widetilde {\boldsymbol {r}}_{(i)}\right )^{-1}\widetilde {\boldsymbol {r}}_{(i)}^{T}\boldsymbol {C}_{(i)}^{-1}\) is the i th “block-row” of C− 1. Note, this is really a Ni × (Ntot − Ni) matrix, where Ntot is the total number of data points, but we refer to it as a “block-row” because of its association with the i th rows of the correlation matrix. This quantity will be noted \(\boldsymbol {C}_{i,(i)}^{-1}\), e.g., the i th block-row of C− 1 not including the i th block-diagonal portion. Therefore,
The advantage of these manipulations will become clear when returning to Eqs. C.15 and C.16 as follows:
In the above manipulations μ(xi) = μi as follows: \(\left [\begin {array}{ccc}\boldsymbol {C}_{i,(i)}^{-1} & \boldsymbol {C}_{ii}^{-1} \end {array}\right ]=\boldsymbol {C}^{-1}_{i}\) e.g. the ith block-row, and \(\left (\boldsymbol {C}_{ii}^{-1}\right )^{-1} \boldsymbol {C}_{ii}^{-1}=\boldsymbol {I}\) where I is the identity matrix. These manipulations enable the direct computation of the leave-i-out cross-validation. Computationally, this is a much more favorable estimate over the alternative which would require retraining many times for however many i there are. This methodology can easily be applied towards k-folds cross-validation where the previously introduced i would correspond indices in the matrix belonging to each of the k-folds.
An expression for the leave-one-out prediction variance can be obtained using the same matrix manipulations. For i th hold out case, the prediction covariance for \(\hat {\boldsymbol {f}}_{(i)}(\boldsymbol {x}_{i})\) can be obtained from the following:
the diagonal of which contains the prediction variance estimates.
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fernandez-Zelaia, P., Melkote, S.N. Process-Structure-Property Modeling for Severe Plastic Deformation Processes Using Orientation Imaging Microscopy and Data-Driven Techniques. Integr Mater Manuf Innov 8, 17–36 (2019). https://doi.org/10.1007/s40192-019-00125-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-019-00125-8