Abstract
Introduction
The randomized, placebo-controlled, double-blind MOVe-OUT trial demonstrated molnupiravir (800 mg every 12 h for 5 days) as safe and effective for outpatient treatment of mild-to-moderate COVID-19, significantly reducing the risk of hospitalization/death in high-risk adults. At the time of that report, virologic assessments from the trial were partially incomplete as a result of their time-intensive nature. Here we present final results from all prespecified virology endpoints in MOVe-OUT based on the full trial dataset.
Methods
Nasopharyngeal swabs were collected at baseline (day 1, prior to first dose) and days 3, 5 (end-of-treatment visit), 10, 15, and 29. From these samples, change from baseline in SARS-CoV-2 RNA titers (determined by quantitative PCR), detection of infectious SARS-CoV-2 (by plaque assay), and SARS-CoV-2 viral error induction (determined by whole genome next-generation sequencing) were assessed as exploratory endpoints.
Results
Molnupiravir was associated with greater mean reductions from baseline in SARS-CoV-2 RNA than placebo (including 50% relative reduction at end-of-treatment) through day 10. Among participants with infectious virus detected at baseline (n = 96 molnupiravir, n = 97 placebo) and evaluable post-baseline samples, no molnupiravir-treated participant had infectious SARS-CoV-2 by day 3, whereas infectious virus was recovered from 21% of placebo-arm participants on day 3 and 2% at end-of-treatment. Consistent with molnupiravir’s mechanism of action, sequence analysis demonstrated that molnupiravir was associated with an increased number of low-frequency transition errors randomly distributed across the SARS-CoV-2 RNA genome compared with placebo (median 143.5 molnupiravir, 15 placebo), while transversion errors were infrequent overall (median 2 in both arms). Outcomes were consistent regardless of baseline SARS-CoV-2 clade, presence of SARS-CoV-2-specific immune response, or viral load.
Conclusions
A 5-day course of orally administered molnupiravir demonstrated a consistently greater virologic effect than placebo, including rapidly eliminating infectious SARS-CoV-2, in high-risk outpatients with mild-to-moderate COVID-19.
Trial Registration
ClinicalTrials.gov, NCT04575597.
Similar content being viewed by others
Why carry out the study? |
MOVe-OUT was a randomized, placebo-controlled, double-blind phase 2/3 trial that demonstrated a 5-day course of molnupiravir as safe and effective for the treatment of mild-to-moderate COVID-19 in non-hospitalized adults at high risk of progression to severe disease. When these safety and efficacy data were published, the much more time-intensive virologic assessments from the trial had been only partially completed. |
Here we report final results from all prespecified, exploratory virology endpoints from MOVe-OUT based on the full trial dataset, in order to compare the virologic effects of molnupiravir and placebo in this patient population. |
What was learned from the study? |
Molnupiravir was associated with a 50% relative reduction in SARS-CoV-2 RNA compared to placebo at the end of treatment. |
Molnupiravir rapidly eliminated infectious SARS-CoV-2. |
A 5-day course of orally administered molnupiravir demonstrated consistently greater virologic effect than placebo regardless of baseline SARS-CoV-2 clade, presence of SARS-CoV-2-specific immune response, or viral load. |
Introduction
The COVID-19 pandemic, caused by the SARS-CoV-2 coronavirus, has caused seven million deaths worldwide since 2019 [1]. The continued emergence of new SARS-CoV-2 variants and waning humoral immunity after prior infection and/or vaccination are of major concern [2,3,4,5,6,7]. Regardless of vaccination status, reinfection with SARS-CoV-2 carries a significant risk of hospitalization and death in high-risk individuals [8]. Therefore, easily self-administered antiviral therapies that reduce the likelihood of COVID-19 progression remain important to the public health response [9, 10]. Molnupiravir is a small-molecule ribonucleoside prodrug of N-hydroxycytidine (NHC) with broad antiviral activity against RNA viruses, including SARS-CoV-2 and its variants (e.g., Omicron and Delta) [11,12,13,14,15,16,17,18,19,20], and a high barrier to resistance development [11, 13, 21,22,23]. After oral administration of molnupiravir, NHC circulates systemically before undergoing intracellular phosphorylation to NHC triphosphate. NHC triphosphate can substitute for the ribonucleotides CTP and UTP during viral replication, which causes an accumulation of deleterious errors throughout the viral genome, ultimately reducing viral infectivity and replication [13, 22, 24,25,26].
In the MOVe-OUT trial, molnupiravir was superior to placebo in reducing the risk of all-cause hospitalization/death through day 29 in adults with mild-to-moderate COVID-19 and risk factors for severe illness [27]. Molnupiravir also improved secondary clinical outcomes, including more rapid symptom resolution, faster normalization of COVID-19 biomarkers, less need for respiratory interventions, and shorter length of hospital stay if hospitalization was required [27, 28]. When the clinical efficacy data from MOVe-OUT were first reported, some virologic assessments were still incomplete as a result of the time-intensive nature and low throughput of the prerequisite laboratory assays [27]. This paper reports final results from all prespecified, exploratory virology endpoints based on the full trial dataset, in order to compare the virologic effects of molnupiravir and placebo in non-hospitalized, high-risk adults with mild-to-moderate COVID-19.
Methods
Trial Design
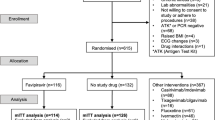
Here we report additional data from the phase 3 component of MOVe-OUT (Protocol MK4482-002), a randomized, placebo-controlled, double-blind, parallel-group, phase 2/3 trial evaluating molnupiravir for COVID-19 (clinicaltrials.gov NCT04575597), which was previously described in detail [27, 29]. The full trial protocol is included as Supplementary Material online.
Eligible participants for this trial were non-vaccinated, non-hospitalized adults with mild-to-moderate COVID-19 (sign/symptom onset ≤ 5 days prior to randomization), laboratory-confirmed SARS-CoV-2 infection ≤ 5 days prior, and ≥ 1 risk factor for developing severe illness from COVID-19 (> 60 years old, active cancer, chronic kidney disease, chronic obstructive pulmonary disease, diabetes mellitus, obesity, and/or serious heart conditions). Participants were randomized to orally administered molnupiravir (800 mg, twice daily, for 5 days) or matched placebo. Other drugs and biologics intended as COVID-19 treatments, except antipyretics, anti-inflammatories, and/or corticosteroids, were prohibited.
Trial Ethics
The trial was conducted in accordance with principles of Good Clinical Practice and was approved by the appropriate institutional review boards/ethics committees and regulatory agencies. Written informed consent was provided by all participants prior to their enrollment into the trial. All authors made substantial contributions to the conception/design of the work and/or the acquisition, analysis, or interpretation of data for the work; either drafted the manuscript or reviewed it critically for important intellectual content; and approved the final version of the manuscript for publication.
Methodology of Virologic Assessments
Serum SARS-CoV-2 anti-nucleocapsid antibodies were assessed at a central laboratory using the Elecsys® assay (Roche Diagnostics, Indianapolis, IN, USA) and serum SARS-CoV-2 anti-spike neutralizing antibodies were assessed using the SARS-CoV-2 PhenoSense® nAB assay (Monogram Biosciences, South San Francisco, CA, USA), at baseline, day 5 (anti-nucleocapsid antibodies only), day 10, and day 29. Nasopharyngeal swabs were collected from participants at baseline (day 1, prior to first dose) and days 3, 5 (end-of-treatment visit), 10, 15, and 29. SARS-CoV-2 testing performed for eligibility screening was conducted using local standard-of-care assays prior to enrollment. All subsequent virologic assessments were conducted at a central laboratory.
Viral RNA
SARS-CoV-2 RNA titers from each nasopharyngeal sample were evaluated by reverse transcriptase polymerase chain reaction (RT-PCR), using two types of assays: a quantitative assay (lower limit of quantitation 500 copies SARS-CoV-2 RNA/mL) and qualitative assay (lower limit of detection 1800 nucleic acid amplification test detectable units/mL; cycle threshold [Ct] value ≤ 35). The research-use-only quantitative assay was developed at Q2 Solutions (Morrisville, NC, USA). After RNA extraction from nasopharyngeal samples using the MagMAX™ viral/pathogen nucleic acid isolation kit on the KingFisher™ sample purification system (ThermoFisher Scientific, Waltham, MA, USA), samples underwent RT-PCR and quantification using the TaqPath™ RT-PCR COVID-19 Kit on either the 7500 Fast Real-Time PCR or the 7500 Fast Dx Real-Time systems (ThermoFisher Scientific). The qualitative assay was the commercially available Cobas® SARS-CoV-2 assay (Roche Diagnostics, Indianapolis, IN, USA) as validated under emergency use authorization.
Infectivity
Given the low probability of detecting cell-culture infectious virus in patient samples with low viral RNA titers, specifically titers less than 106 SARS-CoV-2 RNA copies/mL [30,31,32,33], only those samples with ≥ 100,000 SARS-CoV-2 RNA copies/mL were assessed for infectious SARS-CoV-2. Infectivity was assessed using an exploratory, first-generation plaque assay. For this assay, samples were serially diluted in duplicate in serum-free Eagle’s minimum essential medium; 100-μL aliquots of diluted samples were placed in 24-well plates pre-seeded with > 90% confluent Vero E6 cells and incubated for 60 min at 37 °C and 5% CO2, before being topped with 1 mL of overlay medium and incubated for 48 h. Plaques were subsequently visualized by crystal violet staining and manually counted. The lower limit of quantification for this assay was 200 plaque-forming units (PFU)/mL. Titers for both of the duplicate samples were averaged and reported as a single value; average values below 200 were reported as < 200 PFU/mL.
Next-Generation Sequencing (NGS)
Baseline and day 5 nasopharyngeal samples with ≥ 600 SARS-CoV-2 RNA copies/mL underwent whole-genome NGS. In addition, day 10, 15, and 29 samples with high viral RNA titers (≥ 100,000 SARS-CoV-2 RNA copies/mL) also underwent whole-genome NGS based on the possibility that high titers post treatment could potentially be indicative of molnupiravir resistance mutations. In addition, NGS data were also used for viral genotyping and to assess viral error induction further to molnupiravir’s mechanism of action.
Sequencing was performed using the Ion AmpliSeq SARS-CoV-2 research panel (ThermoFisher Scientific, Waltham, MA, USA), which provides > 99% genome coverage. Reverse transcription of input viral RNA and subsequent library preparation were performed using the SuperScript VILO cDNA Synthesis and Ion AmpliSeq Library kits, respectively (ThermoFisher Scientific). After PCR amplification, libraries were partially digested and the final library quantified using the Ion Library TaqMan kit, to inform final library loading onto an Ion Torrent sequencer Ion 540 chip (ThermoFisher Scientific), with a minimum of 250,000 reads per sample. Sequencing data were analyzed using Torrent Suite Software V5.16 (ThermoFisher) by aligning reads to the SARS-CoV-2 reference genome NCBI Genbank Entry MN908947.3 (positions 43–29,842). The assay-validated lower allele frequency detection limit for single nucleotide variants was 18%. Each sample’s viral genome consensus sequence (including detected sequence variations) was assembled using the IRMA report plug-in. The detailed NGS methodology is described in the Supplementary Material online (Methods S1).
On the basis of the NGS data, baseline SARS-CoV-2 clades were assigned with the nextclade tool using each sample’s genome consensus sequence [34]. If baseline NGS data were not available for viral genotyping, clade assignment was made using the next post-baseline sample with available NGS data. The accumulation of low-frequency SARS-CoV-2 RNA errors (a measure that reflects the antiviral mechanism of molnupiravir) within each sample was measured from raw sequencing data; such errors were defined as nucleotide changes with frequencies between 0.4% and 10% of the total number of sample sequence reads compared with the sample consensus sequence. Treatment-emergent amino acid substitutions, defined as those occurring in post-baseline samples from ≥ 2 participants with an allele frequency of ≥ 18% reference genome-aligned NGS reads, were also determined.
Virologic Outcomes Evaluated
The MOVe-OUT trial included several prespecified exploratory endpoints and subgroup analyses, which were based on data from the virologic assessments described above. All of these outcomes were assessed in the modified intention-to-treat (MITT) population (all randomized participants who received ≥ 1 dose of study intervention and were not hospitalized before the first dose).
The endpoint of all-cause hospitalization or death through day 29 was assessed according to causative SARS-CoV-2 clade, baseline anti-SARS-CoV-2 antibody status, and baseline viral load; these subgroup analyses were prespecified in the statistical analysis plan without adjustments for multiple comparisons. In addition, the following exploratory virology endpoints were prespecified: (a) changes in SARS-CoV-2 viral load, expressed as viral RNA titers, from baseline; (b) participants with undetectable SARS-CoV-2 RNA; (c) participants with undetectable infectious SARS-CoV-2; and (d) changes in SARS-CoV-2 RNA sequence from baseline.
Changes in log10 viral load from baseline were estimated using constrained longitudinal analysis models, allowing adjustment for differences in mean baseline viral load and accounting for missing data. Since all of these were exploratory endpoints, none of them underwent formal evaluation via hypothesis testing. All analyses were conducted using SAS version 9.4 (SAS Institute, Cary, NC, USA).
Results
Baseline Virology and Serology
At baseline, SARS-CoV-2 RNA was confirmed in nasopharyngeal samples from 1277/1408 (90.7%) MITT participants through the qualitative assay at the central laboratory (Table 1); since all participants had a positive study-qualifying SARS-CoV-2 test performed locally during screening, the discrepancy was possibly due to sampling variability, false positive local results, and/or differences in assay sensitivities. At baseline, SARS-CoV-2 nucleocapsid antibodies and neutralizing antibodies (suggesting previous SARS-CoV-2 infection or early immune response to infection) were detected in 19.6% and 26.8% of participants, respectively, in the molnupiravir versus 21.6% and 26.9% in the placebo arm (Table 2).
Overall, 1063/1408 (75.5%) of MITT participants had evaluable sequence data for determination of infecting SARS-CoV-2 clade; reasons for unevaluable sequences included missing samples, poor sequence quality because of too low or too high viral RNA titers, or low sequence coverage. The most commonly detected clades were 21J (Delta; 42.1%), 21H (Mu; 12.4%), 21I (Delta; 7.0%), and 20J (Gamma; 6.2%); clade distribution was generally comparable between arms (Table 3).
Clinical Efficacy by Baseline Clade, Serostatus, and Viral Load
All-cause hospitalization or death through day 29 was more common with placebo than molnupiravir for the Mu and Gamma variants and comparable between arms for Delta (comprising clades 21A, 21I, and 21J) (Fig. 1, Table S1). In participants without anti-SARS-CoV-2 nucleocapsid or anti-SARS-CoV-2 neutralizing antibodies at baseline and those participants who had high baseline SARS-CoV-2 viral load, hospitalization/death rates were lower with molnupiravir than placebo; for all other subgroups, hospitalization/death rates were comparable between study arms (Fig. 1, Table S1).
Incidence of hospitalization or death through day 29 by baseline variant, anti-SARS-CoV-2 antibodies, and viral load in the final analysis of the modified intention-to-treat population. CI confidence interval. *These data are based on the final trial dataset and are thus different from the partial results previously reported in [27]. †Outcomes by individual clade are reported in Table S3. ‡Missing data, invalid samples, tests not done, or results reported as “unknown” were all categorized as unknown. Results for this category are reported in Table S3. All subgroups shown were prospectively defined. §High viral load was defined as > 106 copies/mL of SARS-CoV-2 RNA. ||Low viral load was defined as between 500 and > 106 copies/mL of SARS-CoV-2 RNA. ¶Undetectable viral load was defined as < 500 copies/mL of SARS-CoV-2 RNA
Viral Load Changes Over Time
In the quantitative assay, 1211/1408 (86.0%) participants had SARS-CoV-2 RNA detected in baseline nasopharyngeal swabs (Table 1) and were thus included in the longitudinal analysis of changes in viral load over time. Molnupiravir was associated with greater reductions in SARS-CoV-2 RNA than placebo at days 3, 5, and 10, based on differences in geometric least-squares mean values (Fig. 2a); these results were generally consistent across baseline clades (Fig. 2b–e). Similarly, mean changes in SARS-CoV-2 RNA titer from baseline evaluated on a log10 scale were greater with molnupiravir than placebo through day 10 in the overall MITT population and in all subgroups (including participants with low and high baseline viral load), but the differences in mean change from baseline in RNA titer were comparable between arms at day 15 and at day 29 (Table 4). Molnupiravir and placebo had comparable rates of time to PCR negativity (i.e., undetectable viral load in the qualitative assay) through day 29 in participants with qualitative PCR data available at baseline (Fig. 3, Table S2).
Changes in SARS-CoV-2 RNA titer expressed as geometric least-squares mean values, from nasopharyngeal swabs over time in the modified intention-to-treat population: a overall, b with clade 21J (Delta), c with clade 21I (Delta), d with clade 21H (Mu), and e with clade 20J (Gamma). Data shown are derived from a constrained longitudinal analysis model with RNA titer as the response variable and the following variables as covariates: treatment, study visit, treatment by study visit interaction, and time from symptom onset prior to randomization (≤ 3 days vs. > 3 days) as covariates. *95% confidence interval for difference in geometric least-squares mean values molnupiravir vs. placebo excludes zero
Infectivity
Detection of infectious SARS-CoV-2 in nasopharyngeal swabs by cell culture was correlated with both time from symptom onset and baseline viral load (Fig. S1). Infectious virus was generally not detected in samples with RNA titers < 107 copies/mL, supporting the even lower cutoff of 105 copies/mL that was used to select samples for infectivity testing. In participants with infectious virus detected at baseline, none receiving molnupiravir had infectious SARS-CoV-2 reported by day 3, whereas in the placebo arm infectious virus was recovered from 20/96 (20.8%) at day 3 and 2/89 (2.2%) at the end-of-treatment visit (Fig. 4). In the full MITT population, including those without infectious virus detected at baseline, 3/637 (0.5%) molnupiravir-treated participants and 30/643 (4.7%) of those receiving placebo had infectious virus detected at day 3 and 0/623 (0.0%) and 6/616 (1.0%), respectively, at the end-of-treatment visit (Table S3). One molnupiravir-treated participant had infectious virus detected on day 10. This participant was eligible for the trial on the basis of a positive local SARS-CoV-2 test conducted at screening. However, at the central laboratory, nasopharyngeal samples from this participant subsequently tested positive for human metapneumovirus at baseline and negative for SARS-CoV-2 (using independent qualitative and quantitative PCR assays) at baseline, day 3, and day 5. The participant was first reported PCR-positive for SARS-CoV-2 on day 10 (with a viral load of 1.09 × 106 copies/mL), after already having completed molnupiravir treatment. Of note, this participant was not excluded from any of the analyses presented in this paper. Only two samples, both from the placebo arm, yielded infectious virus more than 10 days after symptom onset (Fig. S1). A post hoc analysis showed that in participants with infectious SARS-CoV-2 detected at baseline, the day 29 hospitalization/death rate was about twice as high with placebo (16/98, 16.3%) as with molnupiravir (8/96, 8.3%), for a numerical treatment difference of − 8.0% (95% confidence interval − 17.7, 1.4). Conversely, in participants who tested negative for baseline infectious virus, day 29 hospitalization/death rates were 47/581 (8.1%) with placebo and 40/588 (6.8%) with molnupiravir (difference − 1.3%; 95% confidence interval − 4.4, 1.7).
Viral Error Induction
As anticipated from its mechanism of action, molnupiravir was associated with increased detection of low-frequency viral RNA errors compared with placebo (Fig. S2). The median (interquartile range) viral RNA error rate with molnupiravir was 9 (5–16) at baseline and 151 (29–389) at day 5 versus 8 (5–14) and 17 (10–45), respectively, with placebo; these differences remained consistent across clades (Table S4). Nucleotide transitions (i.e., C ↔ U, G ↔ A) were the most frequently observed changes (median 143.5 with molnupiravir and 15 with placebo), in particular C-to-U and A-to-G, while the number of transversion errors (i.e., G ↔ U, A ↔ U, C ↔ A, C ↔ G) was low, with a median of 2 in both arms (Fig. 5, Table S5). Nucleotide changes at day 5 were distributed randomly across the entire viral genome, including genes encoding structural proteins (e.g., nucleocapsid) and ORF1a and ORF1b encoding nonstructural proteins required for viral replication (Fig. 6). The incidence of treatment-emergent amino acid changes in SARS-CoV-2 replicase complex proteins was comparable in both arms (Table S6), with no evidence for enrichment of any particular mutation. Specific treatment-emergent amino acid substitutions in SARS-CoV-2 replicase complex proteins and the spike protein are listed in Table S7.
Number of SARS-CoV-2 low-frequency RNA errors detected at baseline and day 5 (end-of-treatment visit) in the modified intention-to-treat population, both a ribonucleotide transitions and b ribonucleotide transversions. N number of participants with evaluable sequencing data available at the specific timepoint, by treatment arm. Low-frequency SARS-CoV-2 RNA errors were defined as nucleotide changes occurring at frequencies between 0.4% and 10% of the total number of sequence reads compared with the sample consensus sequence. Each dot represents an individual participant
Distribution of nucleotide changes (with a variant allele frequency of ≥ 18%) across the SARS-CoV-2 genome at day 5 in the modified intention-to-treat population. N number of participants with paired baseline and day 5 evaluable sequencing data, by treatment arm. Highlighted are the protein coding regions for the SARS-CoV-2 viral replicase complex (13,422 to 19,620) and spike protein (21,463 to 25,384)
Discussion
These full virology data from pre-specified, exploratory analyses of the MOVe-OUT trial confirm the antiviral efficacy of molnupiravir against SARS-CoV-2 in patients with COVID-19 and are congruent with the previously reported partial virology results from this trial [27], as well as with corresponding observations from other clinical trials [35,36,37]. Molnupiravir consistently led to rapid reduction of infectious SARS-CoV-2. In participants with infectious SARS-CoV-2 isolated at baseline, none receiving molnupiravir subsequently had detectable infectious virus by day 3, whereas infectious SARS-CoV-2 was recovered from some placebo-arm participants up to the end-of-treatment visit. Molnupiravir also showed greater viral RNA reductions than placebo during the early viral replication period, up to study day 10. This finding is important, because SARS-CoV-2 loads at day 5 and day 10 post treatment initiation were previously identified as strong predictors of clinical outcomes in this high-risk patient population [38]. High SARS-CoV-2 RNA titers persisting through days 5 to 10 despite antiviral treatment are associated with an increased risk of hospitalization/death, as well as an increased probability of requiring mechanical ventilation or supplemental oxygen [38]. At later timepoints, on the other hand, viral RNA titers are of limited clinical relevance and generally due to prolonged shedding of RNA fragments unlikely to be associated with infectious virus [32, 33, 39,40,41]. Even in patients with persistently high viral load, infectious virus is generally not detected after day 10 of COVID-19 symptom onset [32, 40, 42, 43], and there is no known threshold SARS-CoV-2 RNA titer at later timepoints that suggests the presence of infectious virus. In our trial, SARS-CoV-2 RNA titers at day 15 and day 29 were low in both study arms (since viral loads naturally decline over time in immunocompetent patients) and were more comparable between arms at those timepoints. Of note, viral infectivity was not evaluated in samples with low RNA titers of < 105 copies/mL; this threshold was predefined on the basis of published data indicating that even with higher titers, infectious SARS-CoV-2 is infrequently isolated from samples with < 106 copies/mL [30, 33], something that we also observed in our own data. Overall, the virologic response to molnupiravir was consistent, irrespective of baseline viral load, presence of baseline SARS-CoV-2 antibodies baseline (indicating a humoral immune response against the virus), or SARS-CoV-2 clade.
In this updated, full virology dataset from MOVe-OUT, about two-thirds of participants had Delta variant sublineages confirmed by NGS, consistent with the increasing global prevalence of Delta during later phases of participant enrollment into MOVe-OUT (i.e., July–October 2021). Given the timing of the trial, the Omicron variant was not detected in any participant. However, data from in vitro studies confirmed that molnupiravir remains active against all Omicron sublineages evaluated to date [18, 44]. When the MOVe-OUT primary endpoint of all-cause hospitalization or death through day 29 was compared by baseline viral clade, molnupiravir generally performed better than placebo, especially among participants infected with Gamma and Mu. Only for the Delta variant was the treatment effect comparable between arms, driven by the markedly lower rate of hospitalization or death observed in the placebo arm among participants with clade 21J (Delta) compared with other common clades, i.e., 8% versus 16–20%, respectively. Of note, molnupiravir clearly maintained antiviral activity in this subgroup of participants with 21J (Delta); when COVID-19 caused by this Delta clade was treated, molnupiravir was associated with a greater reduction in mean change from baseline in SARS-CoV-2 RNA than placebo and also yielded rapid reduction in the number of participants with infectious virus at all post-baseline visits.
Importantly, it does not appear that molnupiravir had reduced clinical efficacy against the no longer circulating Delta variant. Rather, additional analyses of MOVe-OUT trial data suggest that the lower observed effect size with molnupiravir in the final versus the interim analysis coincided with the increasing predominance of Delta but was likely caused by the cumulative effect of minor differences in baseline characteristics known to be prognostic for progression to severe disease. These cumulative differences among the study population increasingly biased outcomes in favor of participants in the placebo arm, especially during the latter parts of the trial, as shown using multivariable logistic regression models [45]. Important examples of such shifts in prognostic baseline factors were greater proportions of participants in the molnupiravir arm ≥ 75 years old and/or with multiple risk factors (both characteristics increase the COVID-19 progression risk) after the interim analysis compared with the interim analysis population; a lower proportion of participants in the placebo arm with moderate COVID-19 (which also increases the risk of progression to severe COVID-19) after the interim analysis; and imbalances favoring placebo in the post-interim analysis cohort that were not present at the interim analysis (i.e., higher proportions of participants with low/undetectable SARS-CoV-2 RNA and/or with anti-SARS-CoV-2 antibodies at baseline, both known to protect from severe disease) [45].
MOVe-OUT is now the second randomized, controlled clinical trial showing rapid decrease of infectious SARS-CoV-2 with molnupiravir as assessed by plaque assay [37], a finding that was also demonstrated in multiple animal models [12,13,14]. Further evidence is needed to assess whether molnupiravir treatment has a clinically relevant impact on SARS-CoV-2 transmission. In MOVe-OUT, only one molnupiravir-treated participant had infectious SARS-CoV-2 isolated after end of treatment, but this participant did not have confirmed SARS-CoV-2 at baseline through day 5. Instead, the participant probably presented with symptoms of a metapneumovirus infection (which was PCR-confirmed at baseline) and a false-positive SARS-CoV-2 PCR result at screening (conducted locally pre-baseline), given that their centrally assessed nasopharyngeal samples did not test positive for SARS-CoV-2 on both qualitative and quantitative assays from baseline until day 10 (and then with a high viral load). The virologic findings observed in MOVe-OUT are also in line with another randomized, placebo-controlled trial, i.e., the AGILE CST-2 phase 2a study (n = 180 participants), which similarly reported greater decreases in viral load at the end of therapy with molnupiravir than placebo [35]. Molnupiravir also exhibited a consistent antiviral effect in the phase 2 component of MOVe-OUT [29].
Molnupiravir does not directly inhibit viral RNA replication by interfering with the activity of the viral polymerase. Instead, its metabolite NHC exerts its antiviral activity via viral error induction, which leads to the production of defective and/or non-infectious virus [12,13,14]. Since non-infectious viral RNA fragments not yet fully cleared from the nasal cavity can result in positive PCR tests, it is not surprising that molnupiravir- and placebo-treated participants had comparable rates of time to PCR negativity when samples were assessed through a sensitive qualitative SARS-CoV-2 PCR assay. The mechanism of viral error induction underlies NHC’s broad activity across SARS-CoV-2 variants (and other RNA viruses) and its demonstrated high barrier to the development of resistance [11,12,13,14,15,16,17,18,19,20,21,22,23]. Sequencing data confirmed that viral error rates across the SARS-CoV-2 genome were higher with molnupiravir than placebo: these nucleotide errors were primarily C-to-U and G-to-A transitions and randomly distributed throughout the viral genome (including across genes encoding for structural and non-structural proteins), as predicted by molnupiravir’s mechanism of action [24, 25]. The same observations were also reported from the AGILE CST-2 trial, in which molnupiravir treatment similarly resulted in a randomly distributed statistically significant increase in the transition: transversion error ratio and was not associated with selection or accumulation of nucleotide errors at specific gene locations, including no apparent induction of potential resistance mutations [36]. In the MOVe-OUT trial, only a small number of molnupiravir-treated participants had treatment-emergent amino acid changes in the viral replicase complex proteins; none of these observed changes (which were likely induced by molnupiravir’s mechanism of action) are currently known to be associated with molnupiravir resistance. Since molnupiravir treatment leads to a rapid reduction in infectious virus, the transmission of virus with treatment-emergent amino acid changes is unlikely.
The data presented here are exclusive to outpatients with mild-to-moderate COVID-19 at antiviral treatment initiation and should not be applied to patients already hospitalized with severe COVID-19, in whom disease progression is largely driven by an overactive host immune response [46]. Patients who progress to severe disease have generally been infected with SARS-CoV-2 for longer than the time window within which initiation of antiviral therapy still confers clinical benefit, and molnupiravir is not indicated in that population. The analyses described in this report also have certain limitations that need to be considered when interpreting the results. First, trial participants had to be unvaccinated, so the virologic impact of molnupiravir in vaccinated patients with breakthrough infections could not be evaluated. On a related note, only a minority of trial participants had serologic evidence of previous COVID-19 infection (which also confers some degree of protective immunity), while pre-existing immunity from vaccination and/or prior infection is now more common worldwide. Second, the occurrence of symptomatic viral rebound was not assessed in the MOVe-OUT trial and is therefore out of scope for this report. Third, the Delta variant, which no longer appears to be circulating, was very prevalent in our trial population. Finally, the first-generation plaque assay (using a single cell type and manual readout) we employed to evaluate SARS-CoV-2 infectivity may have less sensitivity to detect low levels of infectious virus in nasopharyngeal samples than newer infectivity assays based on PCR or immunofluorescence methods. Regardless of this potential limitation (which would have affected both study arms equally), the magnitude of the observed reduction in infectious virus in favor of molnupiravir over placebo was notable, particularly during the first few days after initiation of study intervention. All samples above the prespecified threshold of 105 RNA copies/mL were evaluated for infectivity beyond the 5-day treatment period (i.e., up to study day 29) However, given the potential limitations of the assay, it is possible that we did not detect some cases of low-level infectious SARS-CoV-2 occurring after end of treatment although this is unlikely, especially when considering that about half of the participants in our trial already had 9 or 10 days of COVID-19 symptoms at the end-of-treatment visit and that infectious virus is rarely isolated from nasal swabs more than 10 days after symptom onset [32, 40, 42, 43]. Our results were consistent with those prior reports, with no sample in the molnupiravir arm and only a small number in the placebo arm having infectious virus recovered beyond 10 days post symptom onset. Of note, there is currently no standard approach to measure SARS-CoV-2 infectivity. While there are no direct comparisons of our assay’s sensitivity with that of other methodologies, a phase 2 trial using a PCR-based culture assay (which theoretically may be more sensitive than our method) similarly observed rapid decreases in infectious virus with molnupiravir through the end of 5-day treatment [37], thus lending further support to our results.
Determining virologic outcomes with molnupiravir in the current phase of the pandemic, which is dominated by Omicron sublineages, requires further evaluation, for example through real-world studies. The results from the open-label, randomized, controlled PANORAMIC trial [9], which enrolled over 26,000 participants (about 95% of whom had received ≥ 3 doses of a COVID-19 vaccine) during a period when Omicron had emerged as the predominant variant have recently been reported. PANORAMIC included a virology substudy in which the primary outcome was SARS-CoV-2 viral load on day 7. In the subset of participants from the intensively sampled virology cohort, SARS-CoV-2 RNA levels on day 7 were undetectable in 7/34 participants (21%) in the molnupiravir plus usual care arm compared with 1/39 (3%) in the usual care-only arm. Furthermore, the mean viral RNA load on day 7 was more than tenfold lower with molnupiravir (mean viral RNA load 3.82 log10) than in the control arm (mean viral RNA load 4.93 log10). Other real-world studies conducted in high-risk patients with COVID-19 (including substantial proportions with COVID-19 vaccination and/or prior SARS-CoV-2 infection) during the Omicron era demonstrated that molnupiravir treatment significantly reduced the risk of hospitalization/death [47,48,49] and of post-acute sequelae of SARS-CoV-2 (i.e., “long COVID”) [50] compared to no treatment and that molnupiravir decreased SARS-CoV-2 viral load [51]. These real-world data provide further evidence of molnupiravir’s antiviral activity against SARS-CoV-2 variants (including Omicron) and also illustrate molnupiravir’s clinical benefits for treating breakthrough COVID-19 in vaccinated patients and/or symptomatic reinfection in patients with prior natural immunity to SARS-CoV-2.
Conclusions
A 5-day course of orally administered molnupiravir demonstrated consistently greater virologic effect than placebo in treating COVID-19, across SARS-CoV-2 clades and regardless of whether participants had already mounted a SARS-CoV-2-specific immune response or had high baseline viral load. NGS data from these patients with COVID-19 confirmed molnupiravir’s mechanism of action, viral error induction, as previously predicted from preclinical data. Molnupiravir consistently reduced SARS-CoV-2 RNA titers more than placebo and led to rapid elimination of infectious virus. Whether this finding translates into an earlier reduction in viral transmission from people with SARS-CoV-2 infections requires further clinical evidence.
Data Availability
The data sharing policy, including restrictions, of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA is available at http://engagezone.msd.com/ds_documentation.php. Requests for access to the clinical study data can be submitted through the Engage Zone site or via email to dataaccess@merck.com.
Change history
25 April 2024
A Correction to this paper has been published: https://doi.org/10.1007/s40121-024-00947-w
References
World Health Organization. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 24 Sept 2023.
Perez-Alos L, Armenteros JJA, Madsen JR, et al. Modeling of waning immunity after SARS-CoV-2 vaccination and influencing factors. Nat Commun. 2022;13(1):1614.
Levin EG, Lustig Y, Cohen C, et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N Engl J Med. 2021;385(24):e84.
Ssentongo P, Ssentongo AE, Voleti N, et al. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: a systematic review and meta-analysis. BMC Infect Dis. 2022;22(1):439.
Butt AA, Talisa VB, Shaikh OS, et al. Relative vaccine effectiveness of a SARS-CoV-2 mRNA vaccine booster dose against the Omicron variant. Clin Infect Dis. 2022;75(12):2161–8.
Brown PE, Fu SH, Bansal A, et al. Omicron BA.1/1.1 SARS-CoV-2 infection among vaccinated Canadian adults. N Engl J Med. 2022;386(24):2337–9.
Goldberg Y, Mandel M, Bar-On YM, et al. Protection and waning of natural and hybrid immunity to SARS-CoV-2. N Engl J Med. 2022;386(23):2201–12.
Bowe B, Xie Y, Al-Aly Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat Med. 2022;28(11):2398–405.
Butler CC, Hobbs FDR, Gbinigie OA, et al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet. 2023;401(10373):281–93.
Dal-Re R, Becker SL, Bottieau E, et al. Availability of oral antivirals against SARS-CoV-2 infection and the requirement for an ethical prescribing approach. Lancet Infect Dis. 2022;22(8):e231–8.
Yoon JJ, Toots M, Lee S, et al. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob Agents Chemother. 2018;62(8):e00766-78.
Cox RM, Wolf JD, Plemper RK. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021;6(1):11–8.
Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883.
Wahl A, Gralinski LE, Johnson CE, et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–7.
Abdelnabi R, Foo CS, De Jonghe S, et al. Molnupiravir inhibits the replication of the emerging SARS-CoV-2 variants of concern (VoCs) in a hamster infection model. J Infect Dis. 2021;224(5):749–53.
Vangeel L, Chiu W, De Jonghe S, et al. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res. 2022;198: 105252.
Meng B, Abdullahi A, Ferreira I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022;603(7902):706–14.
Takashita E, Kinoshita N, Yamayoshi S, et al. Efficacy of antibodies and antiviral drugs against Covid-19 Omicron variant. N Engl J Med. 2022;386(10):995–8.
Uraki R, Kiso M, Iida S, et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Nature. 2022;607(7917):119–27.
Rosenke K, Okumura A, Lewis MC, et al. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. JCI Insight. 2022;7(13):e160108.
Agostini ML, Pruijssers AJ, Chappell JD, et al. Small-molecule antiviral beta-d-N(4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J Virol. 2019;93(24):e01348–19.
Urakova N, Kuznetsova V, Crossman DK, et al. β-d-N4-Hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J Virol. 2018;92(3):e01965–17.
Grobler J, Strizki J, Murgolo N, et al. Molnupiravir maintains antiviral activity against SARS-CoV-2 variants in vitro and in early clinical studies. Open Forum Infect Dis. 2021;8(Suppl 1):S373.
Kabinger F, Stiller C, Schmitzova J, et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol. 2021;28:740–6.
Gordon CJ, Tchesnokov EP, Schinazi RF, et al. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem. 2021;297(1):100770.
Malone B, Campbell EA. Molnupiravir: coding for catastrophe. Nat Struct Mol Biol. 2021;28(9):706–8.
Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al. Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients. N Engl J Med. 2022;386(6):509–20.
Johnson MG, Puenpatom A, Moncada PA, et al. Effect of molnupiravir on biomarkers, respiratory interventions, and medical services in COVID-19: a randomized, placebo-controlled trial. Ann Intern Med. 2022;175(8):1126–34.
Caraco Y, Crofoot GE, Moncada PA, et al. Phase 2/3 trial of molnupiravir for treatment of COVID-19 in nonhospitalized adults. NEJM Evid. 2022. https://doi.org/10.1056/EVIDoa2100043.
Sonnleitner ST, Dorighi J, Jansen B, et al. An in vitro model for assessment of SARS-CoV-2 infectivity by defining the correlation between virus isolation and quantitative PCR value: isolation success of SARS-CoV-2 from oropharyngeal swabs correlates negatively with Cq value. Virol J. 2021;18(1):71.
Singh AK, Stellrecht KA, Arunachalam T, et al. Lack of active SARS-CoV-2 virus in a subset of PCR-positive COVID-19 congregate care patients. J Clin Virol. 2021;141:104879.
Jones TC, Biele G, Muhlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021. https://doi.org/10.1126/science.abi5273.
Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–9.
Aksamentov I, Roemer C, Hodcroft EB, et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6(67):3773.
Khoo SH, FitzGerald R, Saunders G, et al. Molnupiravir versus placebo in unvaccinated and vaccinated patients with early SARS-CoV-2 infection in the UK (AGILE CST-2): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Infect Dis. 2023;23(2):183–95.
Donovan-Banfield I, Penrice-Randal R, Goldswain H, et al. Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE phase IIa clinical trial. Nat Commun. 2022;13(1):7284.
Fischer WA 2nd, Eron JJ Jr, Holman W, et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci Transl Med. 2022;14(628):eab17430.
Chawla A, Birger R, Wan H, et al. Factors influencing COVID-19 risk: insights from molnupiravir exposure-response modeling of clinical outcomes. Clin Pharmacol Ther. 2023;113(6):1337–45.
Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–51.
Cevik M, Tate M, Llloyd O, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13–22.
Puhach O, Meyer B, Eckerle I. SARS-CoV-2 viral load and shedding kinetics. Nat Rev Microbiol. 2023;21(3):147–61.
Kang S-W, Park H, Kim JY, et al. Comparison of culture-competent virus shedding duration of SARS-CoV-2 Omicron variant in regard to vaccination status: a prospective cohort study. Vaccine. 2023;41(17):2769–72.
Boucau J, Marino C, Regan J, et al. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N Engl J Med. 2022;387(3):275–7.
Takashita E, Yamayoshi S, Simon V, et al. Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5 subvariants. N Engl J Med. 2022;387(5):468–70.
European Medicines Agency. Withdrawal assessment report for Lagevrio. First published 08 September, 2023. Amsterdam: EMA. 2023. https://www.ema.europa.eu/en/medicines/human/withdrawn-applications/lagevrio. Accessed 24 Sept 2023.
Arribas JR, Bhagani S, Lobo SM, et al. Randomized trial of molnupiravir or placebo in patients hospitalized with Covid-19. NEJM Evid. 2022. https://doi.org/10.1056/EVIDoa2100044.
Xie Y, Bowe B, Al-Aly Z. Molnupiravir and risk of hospital admission or death in adults with Covid-19: emulation of a randomized target trial using electronic health records. BMJ. 2023;380: e072705.
Wong CHK, Au ICH, Lau KTK, et al. Real-world effectiveness of molnupiravir and nirmatrelvir plus ritonavir against mortality, hospitalisation, and in-hospital outcomes among community-dwelling, ambulatory patients with confirmed SARS-CoV-2 infection during the Omicron wave in Hong Kong: an observational study. Lancet. 2022;400(10359):1213–22.
Wai AK, Chan CY, Cheung AW, et al. Association of molnupiravir and nirmatrelvir-ritonavir with preventable mortality, hospital admissions and related avoidable healthcare system cost among high-risk patients with mild to moderate COVID-19. Lancet Reg Health West Pac. 2023;30: 100602.
Xie Y, Choi T, Al-Aly Z. Molnupiravir and risk of post-acute sequelae of Covid-19: cohort study. BMJ. 2023;381:e074572.
Mazzotta V, Lepri AC, Colavita F, et al. Viral load decrease in SARS-CoV-2 BA.1 and BA.2 Omicron sublineages infection after treatment with monoclonal antibodies and direct antiviral agents. J Med Virol. 2023;95(1):e28186.
Acknowledgements
We thank the participants and their families and caregivers for their participation in this trial.
Medical Writing/Editorial Assistance
Medical writing assistance was provided by Dominik J. Wolf, MSc, and editorial assistance was provided by Carol Zecca, BS, both of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA.
Funding
Funding for this research was provided by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA, also funded the rapid service fee for publication.
Author information
Authors and Affiliations
Contributions
Conception or design of the work: JMS, JAG, MGJ, JD, MLB, AP, and CDA. Data collection: JMS, MGJ, PC, and MLB. Data analysis and/or data interpretation: JMS, JAG, NM, AF, MGJ, JD, MLB, AP, and CDA. Drafting the article: JMS and MGJ. Critical revision of the article: All authors. Final approval of the version to be published: All authors.
Corresponding author
Ethics declarations
Conflict of Interest
Julie M. Strizki, Nicholas Murgolo, Arthur Fridman, Matthew G. Johnson, Patricia Carmelitano, Michelle L. Brown, Amanda Paschke, and Carisa De Anda are employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA (MSD), who may own stock and/or hold stock options in Merck & Co., Inc., Rahway, NJ, USA. Jiejun Du and Jay A. Grobler were employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA (MSD), at the time the study was conducted. Jiejun Du is currently an employee of Moderna, and Jay A. Grobler is currently an employee of Pfizer Inc.
Ethical Approval
Our company’s approach to the conduct of clinical trials is in accordance with the ethical principles that have their origin in the Declaration of Helsinki, and that are consistent with Good Clinical Practice and the applicable regulatory requirement(s). The trial was conducted in accordance with principles of Good Clinical Practice and was approved by the appropriate institutional review boards/ethics committees and regulatory agencies at all institutions/study sites (see Table S8). Written informed consent was provided by all participants prior to their enrollment into the trial. No identifying information for any trial participant is included in the manuscript. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship on this publication, take responsibility for the integrity of the work as a whole, and have given their approval for this final version to be published.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Strizki, J.M., Grobler, J.A., Murgolo, N. et al. Virologic Outcomes with Molnupiravir in Non-hospitalized Adult Patients with COVID-19 from the Randomized, Placebo-Controlled MOVe-OUT Trial. Infect Dis Ther 12, 2725–2743 (2023). https://doi.org/10.1007/s40121-023-00891-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-023-00891-1